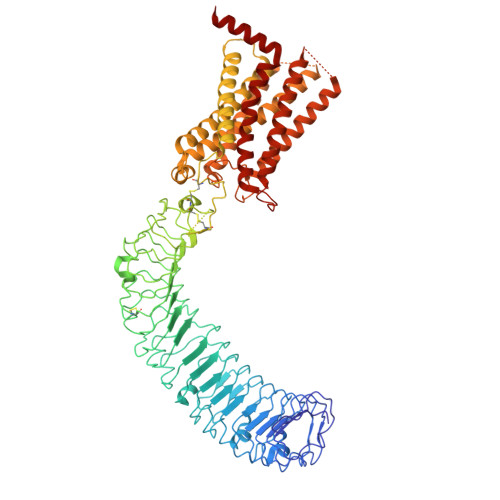
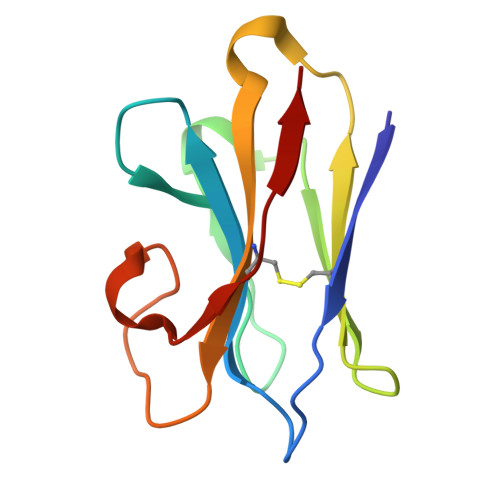
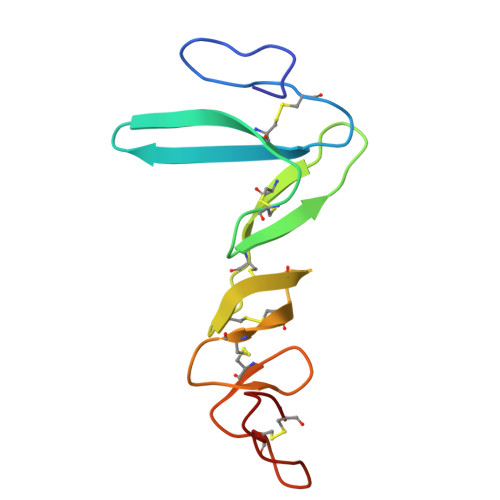
Structural insights into the LGR4-RSPO2-ZNRF3 complexes regulating WNT/ beta-catenin signaling.
Wang, L., Hu, F., Cui, Q., Qiao, H., Li, L., Geng, T., Li, Y., Sun, Z., Zhou, S., Lan, Z., Guo, S., Hu, Y., Wang, J., Yang, Q., Wang, Z., Dai, Y., Geng, Y.(2025) Nat Commun 16: 362-362
- PubMed: 39753551
- DOI: https://doi.org/10.1038/s41467-024-55431-3
- Primary Citation of Related Structures:
8XFP, 8XFS, 8XFT, 8Y69 - PubMed Abstract:
WNT/β-catenin signaling plays key roles in development and cancer 1,2 . ZNRF3/RNF43 modulates Frizzleds through ubiquitination, dampening WNT/β-catenin signaling. Conversely, RSPO1-4 binding to LGR4-6 and ZNRF3/RNF43 enhances WNT/β-catenin signaling 3-5 . Here, we elucidate the overall landscape of architectures in multiple LGR4, RSPO2, and ZNRF3 assemblies, showcasing varying stoichiometries and arrangements. These structures reveal that LGR4 and RSPO2 capture distinct states of ZNRF3. The intrinsic heterogeneity of the LGR4-RSPO2-ZNRF3 assembly is influenced by LGR4 content. Particularly, in the assembly complex with a 2:2:2 ratio, two LGR4 protomers induce and stabilize the inactive state of ZNRF3, characterized by a wide inward-open conformation of two transmembrane helices (TM helices). This specific assembly promotes a stable complex, facilitating LGR4-induced endocytosis of ZNRF3. In contrast, the active dimeric ZNRF3, bound by a single LGR4, adopts a coiled-coil TM helices conformation and dimerization of RING domains. Our findings unveil how LGR4 content mediates diverse assemblies, leading to conformational rearrangements in ZNRF3 to regulate WNT/β-catenin signaling, and provide a structural foundation for drug development targeting Wnt-driven cancers.
- State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: