Structure of Semliki Forest virus in complex with VLDLR
Zheng, T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein E3 | 52 | Semliki Forest virus 4 | Mutation(s): 0 | 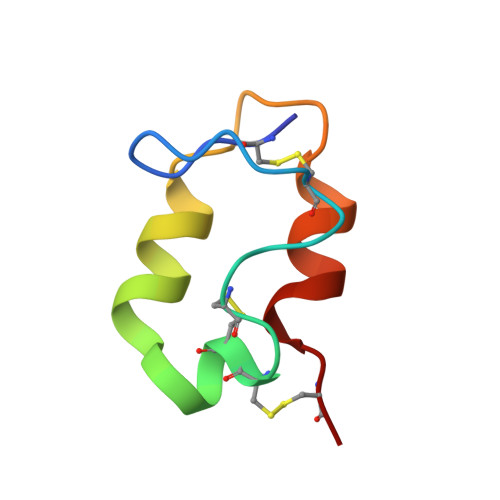 | |
UniProt | |||||
Find proteins for P03315 (Semliki forest virus) Explore P03315 Go to UniProtKB: P03315 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03315 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| capsid protein, partial | 161 | Semliki Forest virus 4 | Mutation(s): 0 | 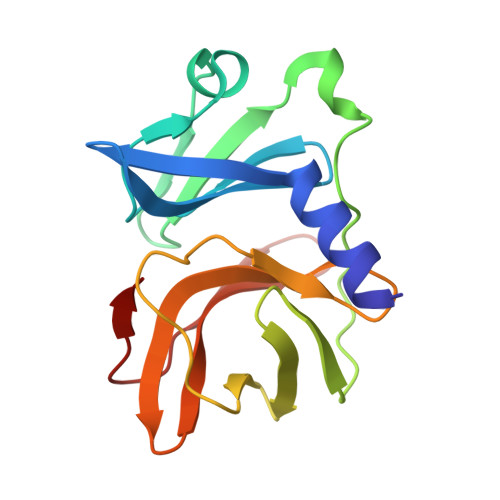 | |
UniProt | |||||
Find proteins for P03315 (Semliki forest virus) Explore P03315 Go to UniProtKB: P03315 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03315 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein E1 | 438 | Semliki Forest virus 4 | Mutation(s): 0 | 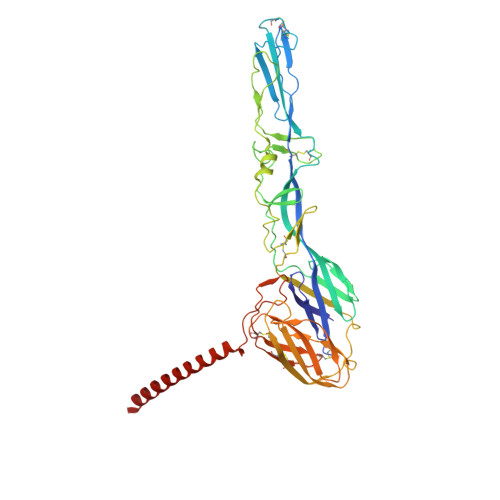 | |
UniProt | |||||
Find proteins for P03315 (Semliki forest virus) Explore P03315 Go to UniProtKB: P03315 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03315 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein E2 | 418 | Semliki Forest virus 4 | Mutation(s): 0 | 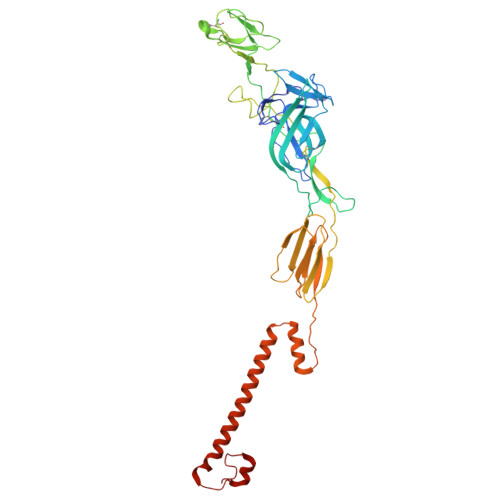 | |
UniProt | |||||
Find proteins for P03315 (Semliki forest virus) Explore P03315 Go to UniProtKB: P03315 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03315 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Very low-density lipoprotein receptor | K [auth A] | 162 | Homo sapiens | Mutation(s): 0 Gene Names: VLDLR | 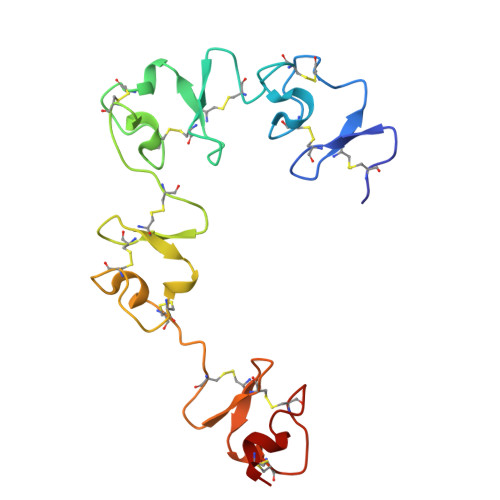 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P98155 (Homo sapiens) Explore P98155 Go to UniProtKB: P98155 | |||||
PHAROS: P98155 GTEx: ENSG00000147852 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P98155 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | AC [auth MA], AD [auth mA], BC [auth NA], BD [auth nA], CC [auth OA], AC [auth MA], AD [auth mA], BC [auth NA], BD [auth nA], CC [auth OA], CD [auth oA], DC [auth PA], DD [auth pA], EC [auth QA], ED [auth qA], FC [auth RA], FD [auth rA], GC [auth SA], GD [auth sA], HC [auth TA], HD [auth tA], IC [auth UA], ID [auth uA], JC [auth VA], KC [auth WA], LC [auth XA], MC [auth YA], NB [auth C], NC [auth ZA], OB [auth k], OC [auth aA], PB [auth BA], PC [auth bA], QB [auth CA], QC [auth cA], RB [auth DA], RC [auth dA], SB [auth EA], SC [auth eA], TB [auth FA], TC [auth fA], UB [auth GA], UC [auth gA], VB [auth HA], VC [auth hA], WB [auth IA], WC [auth iA], XB [auth JA], XC [auth jA], YB [auth KA], YC [auth kA], ZB [auth LA], ZC [auth lA] | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G15407YE GlyCosmos: G15407YE GlyGen: G15407YE | |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |