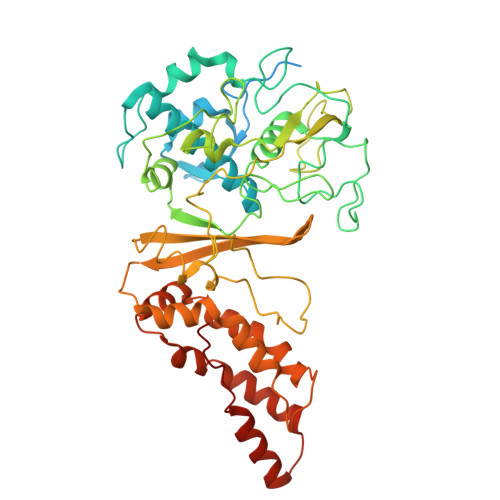
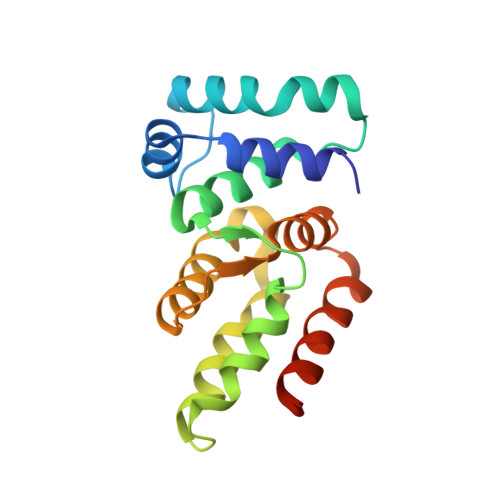
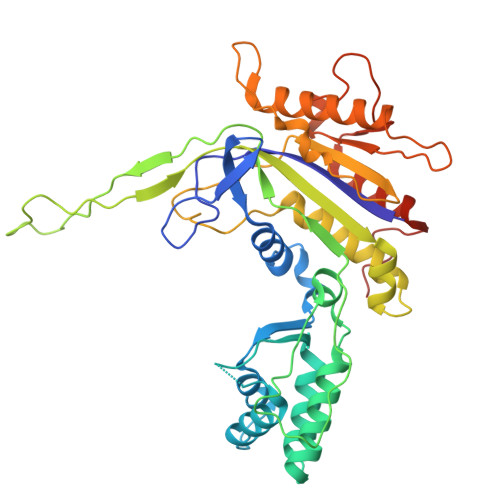
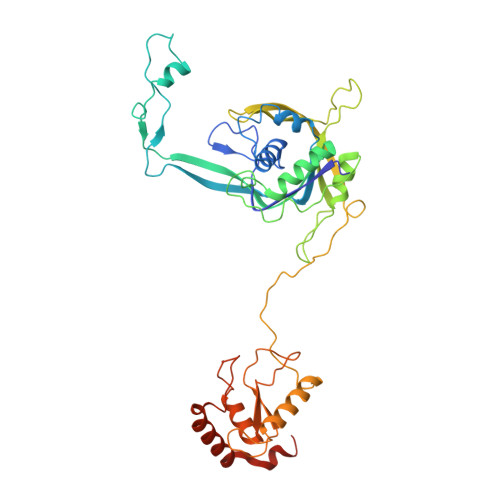
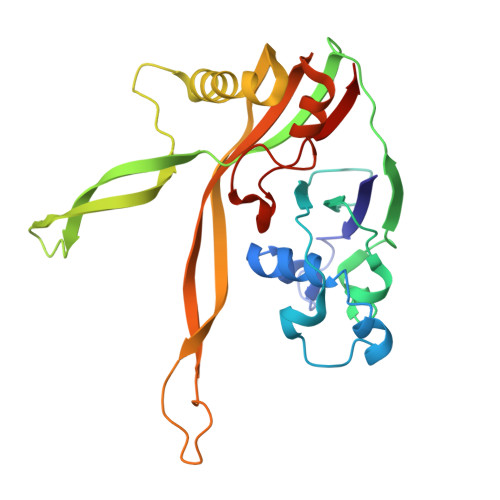
Structural basis for RNA-guided DNA degradation by Cas5-HNH/Cascade complex.
Liu, Y., Wang, L., Zhang, Q., Fu, P., Zhang, L., Yu, Y., Zhang, H., Zhu, H.(2025) Nat Commun 16: 1335-1335
- PubMed: 39904990
- DOI: https://doi.org/10.1038/s41467-024-55716-7
- Primary Citation of Related Structures:
8ZLU, 8ZM3, 8ZOL, 8ZP7, 8ZP9, 9JXS - PubMed Abstract:
Type I-E CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated proteins) system is one of the most extensively studied RNA-guided adaptive immune systems in prokaryotes, providing defense against foreign genetic elements. Unlike the previously characterized Cas3 nuclease, which exhibits progressive DNA cleavage in the typical type I-E system, a recently identified HNH-comprising Cascade system enables precise DNA cleavage. Here, we present several near-atomic cryo-electron microscopy (cryo-EM) structures of the Candidatus Cloacimonetes bacterium Cas5-HNH/Cascade complex, both in its DNA-bound and unbound states. Our analysis reveals extensive interactions between the HNH domain and adjacent subunits, including Cas6 and Cas11, with mutations in these key interactions significantly impairing enzymatic activity. Upon DNA binding, the Cas5-HNH/Cascade complex adopts a more compact conformation, with subunits converging toward the center of nuclease, leading to its activation. Notably, we also find that divalent ions such as zinc, cobalt, and nickel down-regulate enzyme activity by destabilizing the Cascade complex. Together, these findings offer structural insights into the assembly and activation of the Cas5-HNH/Cascade complex.
- Beijing National Laboratory for Condensed Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: