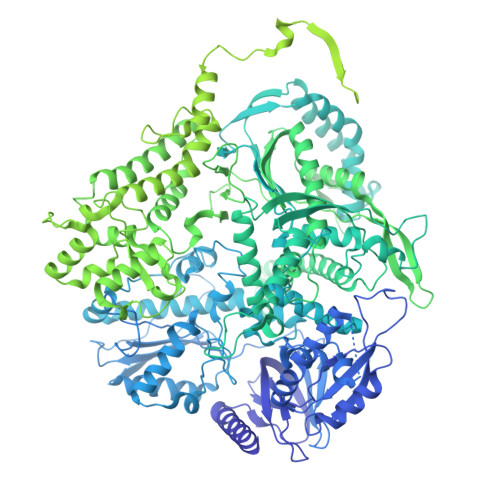
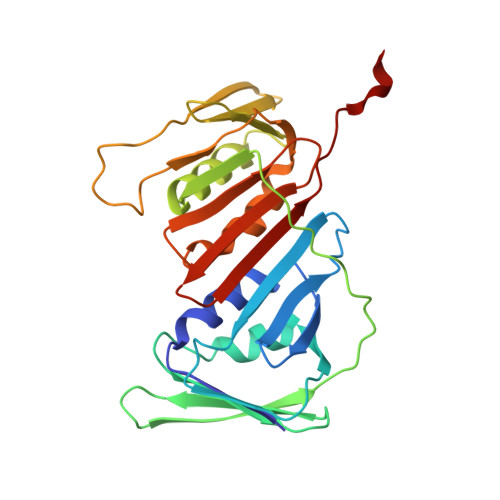
Structures of the human leading strand Pol epsilon-PCNA holoenzyme.
He, Q., Wang, F., Yao, N.Y., O'Donnell, M.E., Li, H.(2024) Nat Commun 15: 7847-7847
- PubMed: 39245668
- DOI: https://doi.org/10.1038/s41467-024-52257-x
- Primary Citation of Related Structures:
9B8S, 9B8T - PubMed Abstract:
In eukaryotes, the leading strand DNA is synthesized by Polε and the lagging strand by Polδ. These replicative polymerases have higher processivity when paired with the DNA clamp PCNA. While the structure of the yeast Polε catalytic domain has been determined, how Polε interacts with PCNA is unknown in any eukaryote, human or yeast. Here we report two cryo-EM structures of human Polε-PCNA-DNA complex, one in an incoming nucleotide bound state and the other in a nucleotide exchange state. The structures reveal an unexpected three-point interface between the Polε catalytic domain and PCNA, with the conserved PIP (PCNA interacting peptide)-motif, the unique P-domain, and the thumb domain each interacting with a different protomer of the PCNA trimer. We propose that the multi-point interface prevents other PIP-containing factors from recruiting to PCNA while PCNA functions with Polε. Comparison of the two states reveals that the finger domain pivots around the [4Fe-4S] cluster-containing tip of the P-domain to regulate nucleotide exchange and incoming nucleotide binding.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI, USA.
Organizational Affiliation: