Rotavirus structure
Jenni, S., Herrmann, T., De Sautu, M., Harrison, S.C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Outer capsid glycoprotein VP7 | 326 | Simian rotavirus A strain RRV | Mutation(s): 0 | 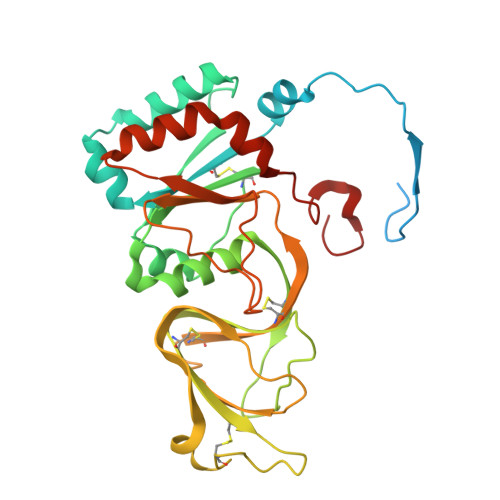 | |
UniProt | |||||
Find proteins for P12476 (Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3])) Explore P12476 Go to UniProtKB: P12476 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12476 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Outer capsid protein VP4 | C [auth 2], D [auth 3], E [auth 4] | 776 | Simian rotavirus A strain RRV | Mutation(s): 0 | 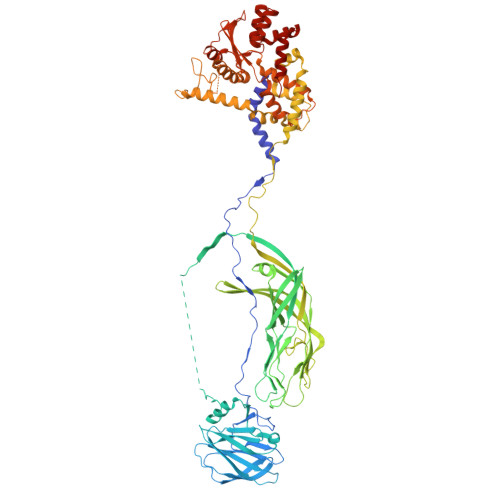 |
UniProt | |||||
Find proteins for P12473 (Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3])) Explore P12473 Go to UniProtKB: P12473 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12473 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Inner capsid protein VP2 | F [auth A], G [auth B] | 887 | Simian rotavirus A strain RRV | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for B3F2X3 (Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3])) Explore B3F2X3 Go to UniProtKB: B3F2X3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B3F2X3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Intermediate capsid protein VP6 | 397 | Simian rotavirus A strain RRV | Mutation(s): 0 | 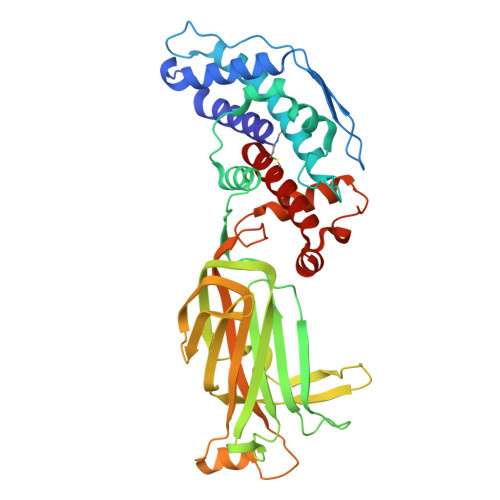 | |
UniProt | |||||
Find proteins for B2BN53 (Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3])) Explore B2BN53 Go to UniProtKB: B2BN53 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B2BN53 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | RA [auth a] | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | CD [auth V] EC [auth Q] EF [auth x] JF [auth y] KE [auth t] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| ZN Query on ZN | AE [auth f] BE [auth f] CB [auth C] CE [auth g] EB [auth C] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CA Query on CA | AB [auth 1] AC [auth P] AD [auth U] AF [auth w] BB [auth 1] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | DB [auth C] DE [auth h] HE [auth j] IB [auth F] OB [auth J] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| FME Query on FME | FA [auth f] GA [auth g] H [auth C] HA [auth h] I [auth D] FA [auth f], GA [auth g], H [auth C], HA [auth h], I [auth D], IA [auth i], J [auth E], JA [auth j], K [auth F], KA [auth k], L [auth G], M [auth H], N [auth I], O [auth J], P [auth K], Q [auth L], R [auth M], S [auth N], T [auth O] | L-PEPTIDE LINKING | C6 H11 N O3 S |  | MET |
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21rc1_5127 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | R01 CA13202 |