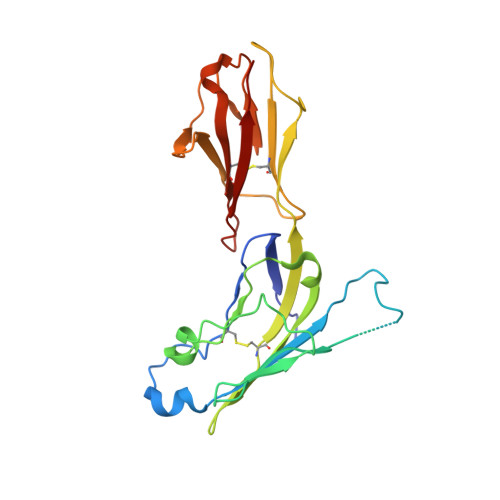
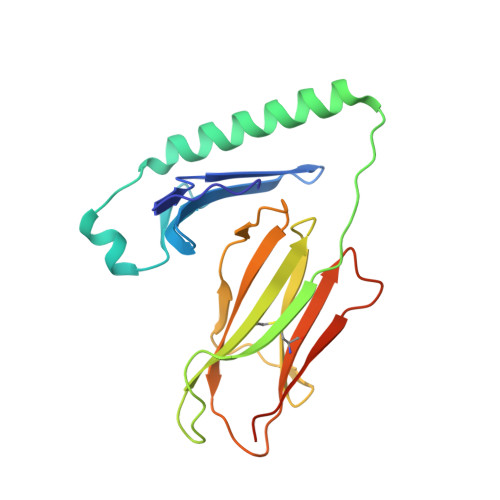
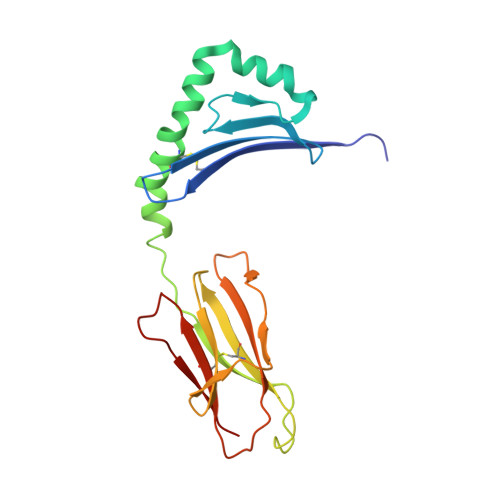
Structural basis for mouse LAG3 interactions with the MHC class II molecule I-A b.
Ming, Q., Antfolk, D., Price, D.A., Manturova, A., Medina, E., Singh, S., Mason, C., Tran, T.H., Smalley, K.S.M., Leung, D.W., Luca, V.C.(2024) Nat Commun 15: 7513-7513
- PubMed: 39209860
- DOI: https://doi.org/10.1038/s41467-024-51930-5
- Primary Citation of Related Structures:
9CYL, 9CYM - PubMed Abstract:
The immune checkpoint protein, Lymphocyte activation gene-3 (LAG3), binds Major Histocompatibility Complex Class II (MHC-II) and suppresses T cell activation. Despite the recent FDA approval of a LAG3 inhibitor for the treatment of melanoma, how LAG3 engages MHC-II on the cell surface remains poorly understood. Here, we determine the 3.84 Å-resolution structure of mouse LAG3 bound to the MHC-II molecule I-A b , revealing that domain 1 (D1) of LAG3 binds a conserved, membrane-proximal region of MHC-II spanning both the α2 and β2 subdomains. LAG3 dimerization restricts the intermolecular spacing of MHC-II molecules, which may attenuate T cell activation by enforcing suboptimal signaling geometry. The LAG3-MHC-II interface overlaps with the MHC-II-binding site of the T cell coreceptor CD4, implicating disruption of CD4-MHC-II interactions as a mechanism for LAG3 immunosuppressive function. Lastly, antibody epitope analysis indicates that multiple LAG3 inhibitors do not recognize the MHC-II-binding interface of LAG3, suggesting a role for functionally distinct mechanisms of LAG3 antagonism in therapeutic development.
Organizational Affiliation:
Moffitt Cancer Center and Research Institute, Department of Immunology, Tampa, FL, 33612, USA.