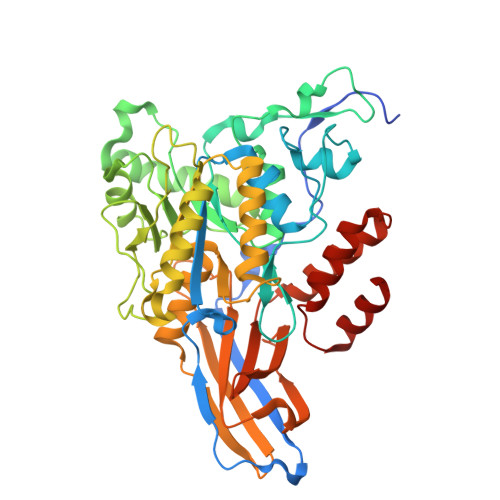
Carbohydrate Deacetylase Unique to Gut Microbe Bacteroides Reveals Atypical Structure.
Schwartz, L.A., Norman, J.O., Hasan, S., Adamek, O.E., Dzuong, E., Lowenstein, J.C., Yost, O.G., Sankaran, B., McLaughlin, K.J.(2025) Biochemistry 64: 180-191
- PubMed: 39663570
- DOI: https://doi.org/10.1021/acs.biochem.4c00519
- Primary Citation of Related Structures:
9D44, 9D4I, 9D5T, 9D60 - PubMed Abstract:
Bacteroides are often the most abundant, commensal species in the gut microbiome of industrialized human populations. One of the most commonly detected species is Bacteroides ovatus . It has been linked to benefits like the suppression of intestinal inflammation but is also correlated with some autoimmune disorders, for example irritable bowel disorder (IBD). Bacterial cell surface carbohydrates, like capsular polysaccharides (CPS), may play a role in modulating these varied host interactions. Recent studies have begun to explore the diversity of CPS loci in Bacteroides ; however, there is still much unknown. Here, we present structural and functional characterization of a putative polysaccharide deacetylase from Bacteroides ovatus ( Bo PDA) encoded in a CPS biosynthetic locus. We solved four high resolution crystal structures (1.36-1.56 Å) of the enzyme bound to divalent cations Co 2+ , Ni 2+ , Cu 2+ , or Zn 2+ and performed carbohydrate binding and deacetylase activity assays. Structural analysis of Bo PDA revealed an atypical domain architecture that is unique to this enzyme, with a carbohydrate esterase 4 (CE4) superfamily catalytic domain inserted into a carbohydrate binding module (CBM). Additionally, Bo PDA lacks the canonical CE4 His-His-Asp metal binding motif and our structures show it utilizes a noncanonical His-Asp dyad to bind metal ions. Bo PDA is the first protein involved in CPS biosynthesis from B. ovatus to be characterized, furthering our understanding of significant biosynthetic processes in this medically relevant gut microbe.
- Department of Chemistry, Vassar College, 124 Raymond Ave, Poughkeepsie, New York 12604, United States.
Organizational Affiliation: