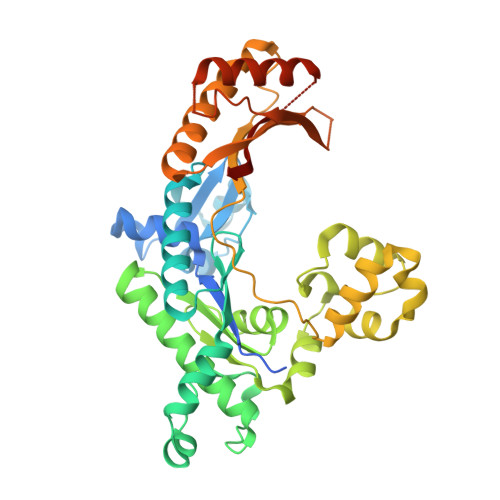
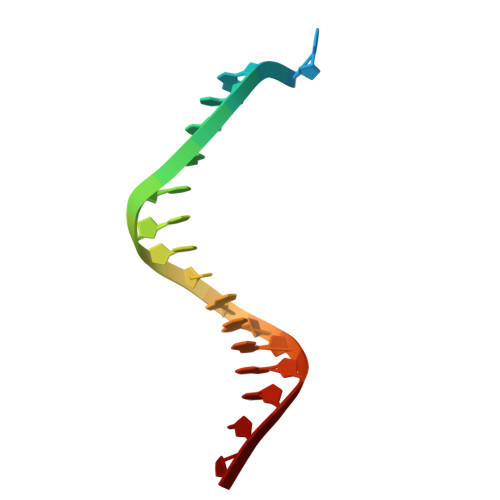
Visualizing DNA polymerase iota catalyze Hoogsteen-directed DNA synthesis.
Frevert, Z., Reusch, D.T., Gildenberg, M.S., Jordan, S.M., Ryan, B.J., Freudenthal, B.D., Washington, M.T.(2025) Nat Commun 16: 5979-5979
- PubMed: 40593703
- DOI: https://doi.org/10.1038/s41467-025-61245-8
- Primary Citation of Related Structures:
9DDR, 9DQT, 9DQU, 9DR7, 9DR9, 9DRB, 9DRC, 9NJH - PubMed Abstract:
Translesion synthesis polymerases efficiently incorporate nucleotides opposite DNA lesions. Pol ι, for example, bypasses minor-groove and exocyclic purine adducts. Conventional X-ray crystallography showed that this enzyme incorporates nucleotides by forming Hoogsteen base pairs with the incoming nucleotide rather than Watson-Crick base pairs. While this revealed the structural basis of nucleotide selection during nucleotide binding, it did not allow the visualization of the process of phosphodiester bond formation or the detection of reaction intermediates that form during nucleotide incorporation. Here, we use a combination of time-lapse crystallography and molecular dynamics simulations to examine the mechanism of pol ι-catalyzed nucleotide incorporation. We show that this enzyme maintains Hoogsteen base pairing with the incoming dNTP during the entire reaction. We also show that pol ι possesses a pyrophosphatase activity that generates two monophosphates within its active site. Our findings provide insights into the features of pol ι's active site that allow it to translocate along DNA and catalyze processive DNA synthesis.
- Department of Biochemistry and Molecular Biology, University of Iowa College of Medicine, Iowa City, IA, USA.
Organizational Affiliation: