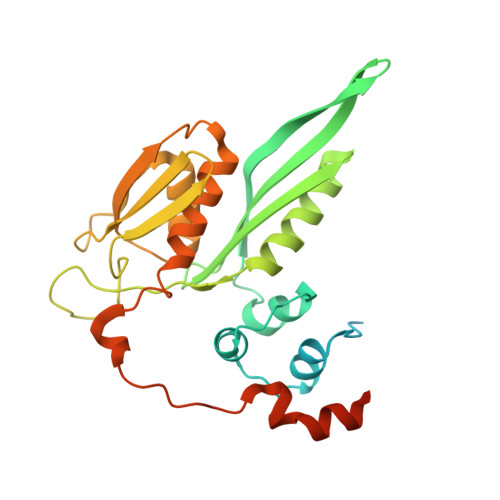
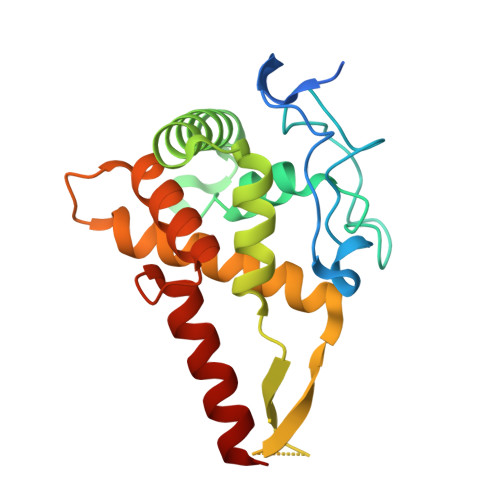
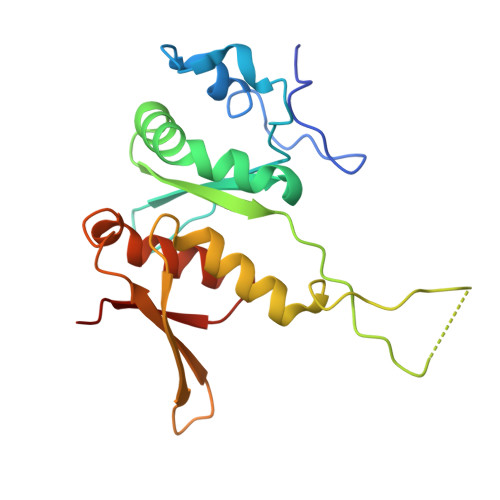
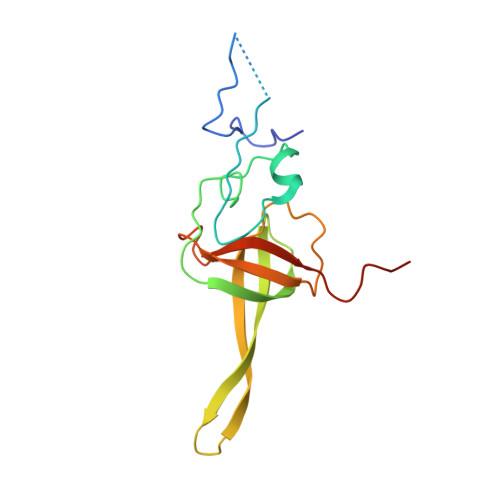
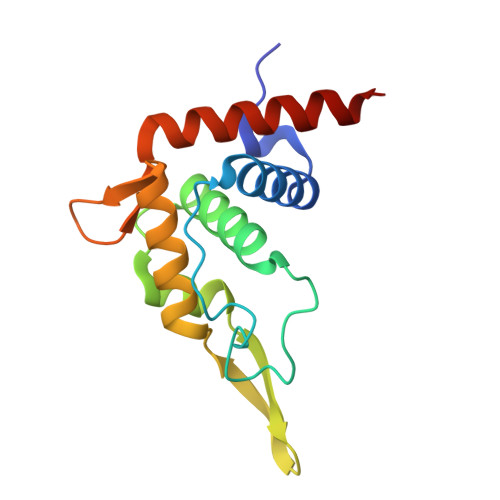
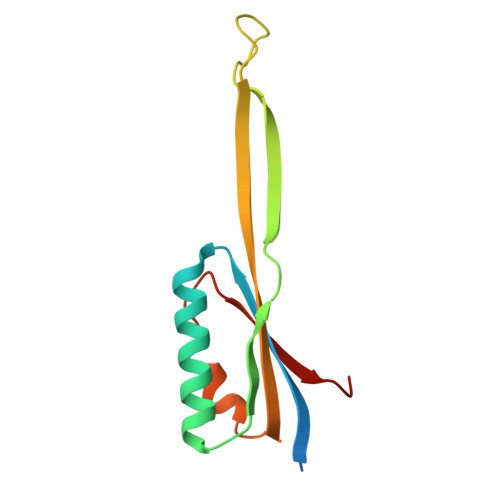
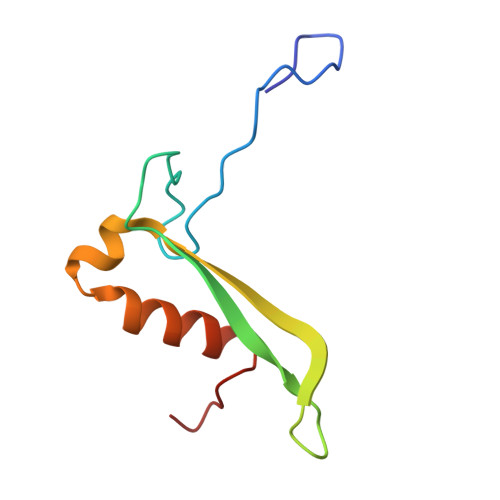
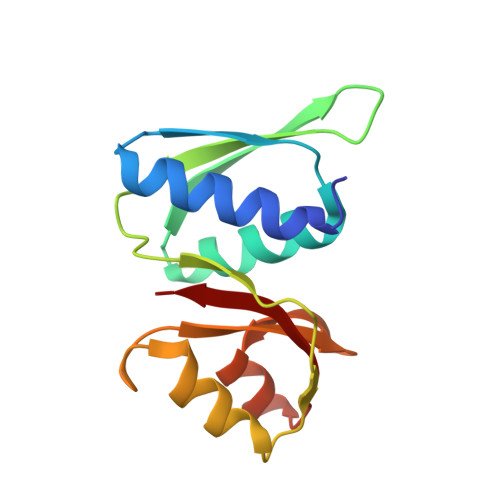
SPIDR enables multiplexed mapping of RNA-protein interactions and uncovers a mechanism for selective translational suppression upon cell stress.
Wolin, E., Guo, J.K., Blanco, M.R., Goronzy, I.N., Gorhe, D., Dong, W., Perez, A.A., Keskin, A., Valenzuela, E., Abdou, A.A., Urbinati, C.R., Kaufhold, R., Rube, H.T., Brito Querido, J., Guttman, M., Jovanovic, M.(2025) Cell 188: 5384-5402.e25
- PubMed: 40701149
- DOI: https://doi.org/10.1016/j.cell.2025.06.042
- Primary Citation of Related Structures:
9ED0 - PubMed Abstract:
RNA-binding proteins (RBPs) regulate all stages of the mRNA life cycle, yet current methods generally map RNA targets of RBPs one protein at a time. To overcome this limitation, we developed SPIDR (split-and-pool identification of RBP targets), a highly multiplexed split-pool method that profiles the binding sites of dozens of RBPs simultaneously. SPIDR identifies precise, single-nucleotide binding sites for diverse classes of RBPs. Using SPIDR, we uncovered an interaction between LARP1 and the 18S rRNA and resolved this interaction to the mRNA entry channel of the 40S ribosome using cryoelectron microscopy (cryo-EM), providing a potential mechanistic explanation for LARP1's role in translational suppression. We explored changes in RBP binding upon mTOR inhibition and identified that 4EBP1 preferentially associates with translationally repressed mRNAs upon mTOR inhibition. SPIDR has the potential to significantly advance our understanding of RNA biology by enabling rapid, de novo discovery of RNA-protein interactions at an unprecedented scale.
- Department of Biological Sciences, Columbia University, New York City, NY 10027, USA.
Organizational Affiliation: