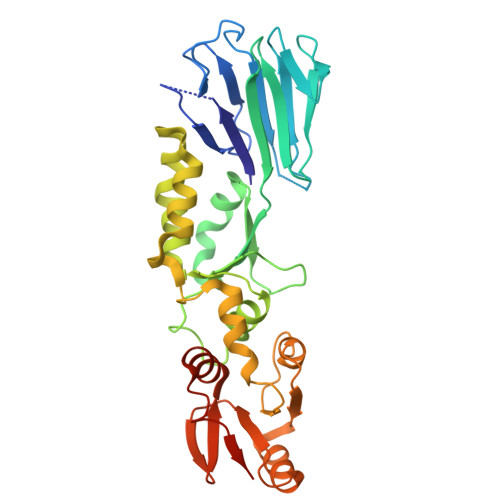
Crystal structure of the folded domains of Xrs2 from Saccharomyces cerevisiae.
Vigneswaran, A., Shi, K., Aihara, H., Evans 3rd, R.L., Latham, M.P.(2025) Acta Crystallogr F Struct Biol Commun 81: 365-373
- PubMed: 40767353
- DOI: https://doi.org/10.1107/S2053230X25006867
- Primary Citation of Related Structures:
9EE7 - PubMed Abstract:
The MRE11-RAD50-NBS1/Xrs2 (MRN/X) protein complex acts as a first responder in DNA double-strand break repair and telomere-length maintenance, yet the structural architecture of the yeast ortholog Xrs2 has remained unresolved. In this study, we present the first structure of the folded N-terminal region of Xrs2 from Saccharomyces cerevisiae, resolved at 2.38 Å using X-ray crystallography. Like the previously determined crystal structures of Schizosaccharomyces pombe Nbs1, the folded structure of S. cerevisiae Xrs2 adopts an extended three-domain organization at its N-terminus. Electrostatic analysis reveals two distinct charged patches: a positively charged patch on the FHA domain and a negatively charged patch in the cleft between the FHA and BRCT1 domains. This charge segregation is likely to play a role in mediating interactions with various ligands.
- Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, MN 55455, USA.
Organizational Affiliation: