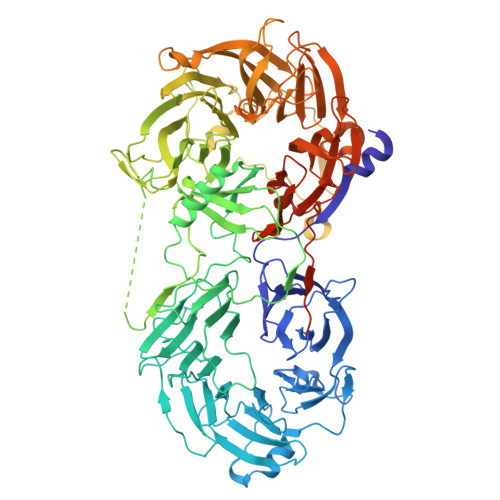
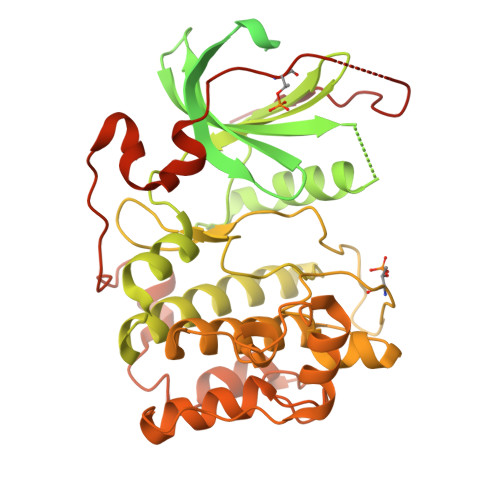
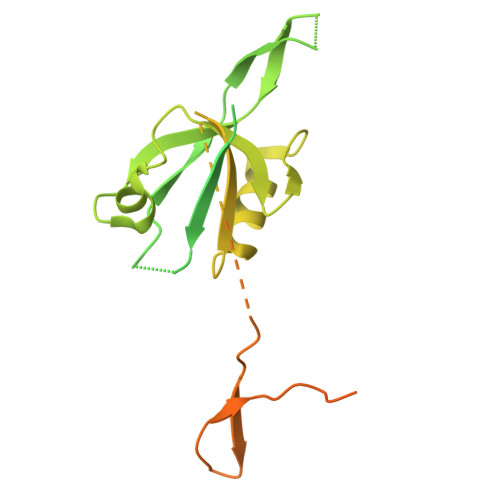
Polarity protein Par6 facilitates the processive phosphorylation of Lgl via a dynamic interaction with aPKC.
Almagor, L., Weis, W.I.(2025) Commun Biol 8: 967-967
- PubMed: 40595028
- DOI: https://doi.org/10.1038/s42003-025-08401-4
- Primary Citation of Related Structures:
9EJK, 9EJL, 9EJM - PubMed Abstract:
Polarity along an apical-basal axis is essential for epithelial cell shape and function. The atypical protein Kinase-C (aPKC) and its regulatory partner Par6 form a complex that is essential for polarization, a primary function of which is to phosphorylate the Lethal giant larvae (Lgl) protein to prevent it from binding to the apical membrane (thereby facilitating its basolateral localization). Par6 binds Lgl directly and is essential for this process, but its mechanism was obscure. Here, we utilize cryo-EM and various biochemical techniques to characterize the interaction of Lgl2 with the aPKCι/Par6 complex and to study the roles of Par6 in promoting Lgl2 phosphorylation. We find that Par6 proteins stabilize a ternary Lgl2/aPKCι/Par6 complex that involves a unique multi-surface interaction of Lgl2 with both aPKCι and Par6. Importantly, we find Par6b induces processive phosphorylation that results in a multi-phosphorylated Lgl2 after a single interaction with the aPKCι/Par6b complex. This is enabled by a Par6b/Lgl2 interaction that maintains contact of Lgl2 with the kinase throughout its distinct nucleotide-binding states. Our results reveal the mechanistic basis for the efficient regulation of Lgl's membrane binding by aPKC/Par6 and provide invaluable structural data for further understanding the mechanisms of this polarity complex.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA, USA. lalmagor@stanford.edu.
Organizational Affiliation: