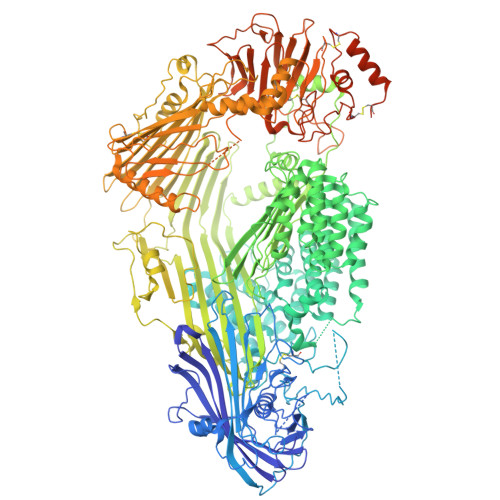
Cryo-EM structure of native honey bee vitellogenin.
Montserrat-Canals, M., Schnelle, K., Leipart, V., Halskau, O., Amdam, G.V., Moeller, A., Cunha, E.S., Luecke, H.(2025) Nat Commun 16: 5736-5736
- PubMed: 40593495
- DOI: https://doi.org/10.1038/s41467-025-58575-y
- Primary Citation of Related Structures:
9ENR, 9ENS - PubMed Abstract:
Vitellogenin (Vg) is the main yolk precursor lipoprotein in almost all egg-laying animals. In addition, along its evolutionary history, Vg has developed a range of new functions in different taxa. In the honey bee, Vg has functions related to immunity, antioxidant protection, social behavior and longevity. However, the molecular mechanisms underlying Vg functionalities are still poorly understood. Here, we report the cryo-EM structure of full-length honey bee Vg, one-step purified directly from hemolymph. The structure provides structural insights into the overall domain architecture, including the lipid binding cavity and the previously uncharacterized von Willebrand factor type D domain. A domain of unknown function has been identified as a C-terminal cystine knot domain based on structural homology. Information about post-translational modifications, cleavage products, metal and lipid binding allow an improved understanding of the mechanisms underlying the range of Vg functionalities. The findings have numerous implications for the structure-function relationship of vitellogenins of other species as well as members of the same protein superfamily, which share the same structural elements.
- Department of Chemistry, University of Oslo, Oslo, Norway.
Organizational Affiliation: