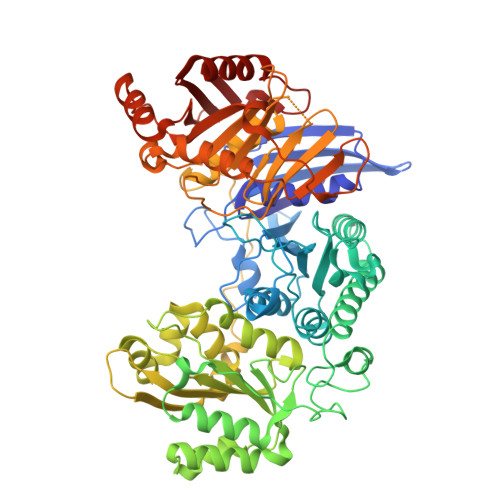
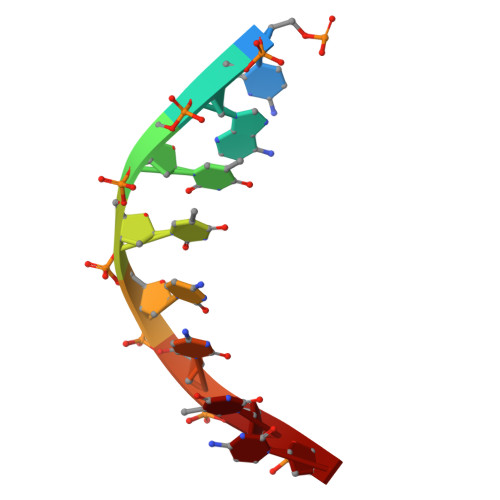
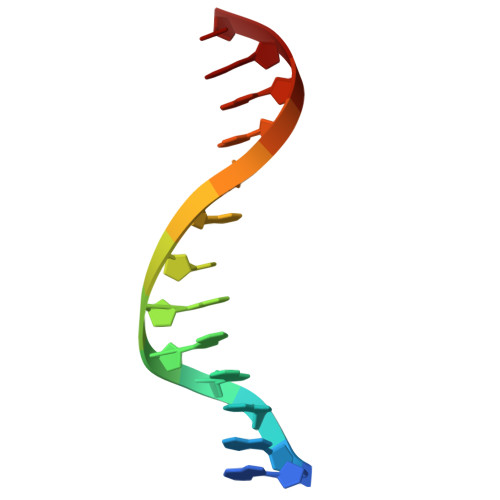
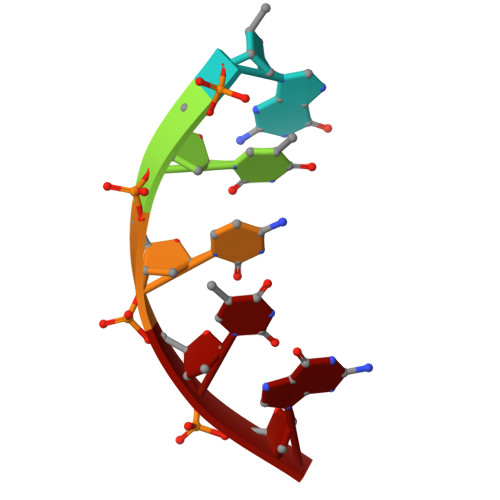
Crystal structures of monomeric BsmI restriction endonuclease reveal coordinated sequential cleavage of two DNA strands.
Sieskind, R., Missoury, S., Madru, C., Commenge, I., Niogret, G., Hollenstein, M., Rondelez, Y., Sauguet, L., Haouz, A., Legrand, P., Delarue, M.(2025) Commun Biol 8: 387-387
- PubMed: 40055548
- DOI: https://doi.org/10.1038/s42003-025-07612-z
- Primary Citation of Related Structures:
9EZ5, 9EZ7, 9EZD, 9F38 - PubMed Abstract:
BsmI, a thermophilic Type IIS restriction endonuclease from Bacillus stearothermophilus, presents a unique structural composition, housing two distinct active sites within a single monomer. Recognition of the non-symmetrical 5'-GAATGC-3' sequence enables precise cleavage of the top and bottom DNA strands. Synthetic biology interventions have led to the transformation of BsmI into Nb.BsmI, a nicking endonuclease. Here we introduce Nt*.BsmI, tailored for top-strand cleavage, which is inactive on standard double-stranded DNA, but active on bottom-strand nicked DNA, suggesting a sequential cleavage mechanism. Crystallographic structures of pre- and post-reactive complexes with cognate DNA show one major conformational change, a retractable loop possibly governing sequential active site accessibility. The x-ray structures reveal the position of the divalent metal ions in the active sites and the DNA:protein interactions, while the models predicted by Alphafold3 are incorrect. This comprehensive structural and functional study lays a foundation for rational enzyme redesign and potential applications in biotechnology.
- Institut Pasteur, Université Paris Cité, CNRS UMR 3528, Unit of Architecture and Dynamics of Biological Macromolecules, 75724, Paris, France.
Organizational Affiliation: