Structural basis for immune cell binding of Fusobacterium nucleatum via the trimeric autotransporter adhesin CbpF.
Marongiu, G.L., Fink, U., Schopf, F., Oder, A., von Kries, J.P., Roderer, D.(2025) Proc Natl Acad Sci U S A 122: e2418155122-e2418155122
- PubMed: 40198705
- DOI: https://doi.org/10.1073/pnas.2418155122
- Primary Citation of Related Structures:
9GH4, 9GH5, 9GH6 - PubMed Abstract:
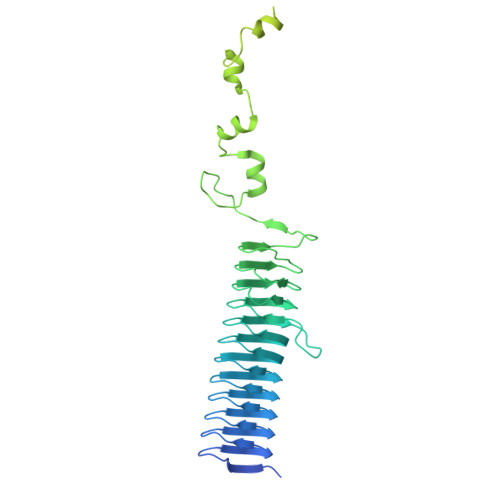
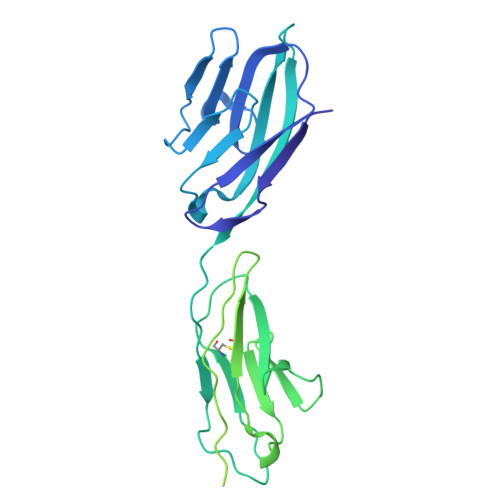
Fusobacterium nucleatum (Fn), a commensal in the human oral cavity, is overrepresented in the colon microbiota of colorectal cancer (CRC) patients and is linked to tumor chemoresistance, metastasis, and a poor therapeutic prognosis. Fn produces numerous adhesins that mediate tumor colonization and downregulation of the host's antitumor immune response. One of these, the trimeric autotransporter adhesin (TAA) CEACAM binding protein of Fusobacterium (CbpF), targets CEACAM1 on T-cells and has been associated with immune evasion of Fn-colonized tumors. Whereas the role of CEACAM1 in homophilic and heterophilic cell interactions and immune evasion is well described, the mechanistic details of its interaction with fusobacterial CbpF remain unknown due to the lack of a high-resolution structure of the adhesin-receptor complex. Here, we present two structures of CbpF alone and in complex with CEACAM1, obtained by cryogenic electron microscopy and single particle analysis. They reveal that CbpF forms a stable homotrimeric complex whose N-terminal part of the extracellular domain comprises a 64 Å long β roll domain with a unique lateral loop extension. CEACAM1 binds to this loop with high affinity via its N-terminal IgV-like domain with a nanomolar dissociation constant as determined by surface plasmon resonance. This study provides a comprehensive structural description of a fusobacterial TAA, illustrates a yet undescribed CEACAM1 binding mode, and paves the way for rational drug design targeting Fn in CRC.
- Leibniz-Forschungsinstitut fur Molekulare Pharmakologie, Berlin 13125, Germany.
Organizational Affiliation: