Revisiting D-Acylases for D-Amino Acid Production.
Martinez-Rodriguez, S., Gavira, J.A.(2025) Microb Biotechnol 18: e70179-e70179
- PubMed: 40491232
- DOI: https://doi.org/10.1111/1751-7915.70179
- Primary Citation of Related Structures:
9G5M, 9GV8 - PubMed Abstract:
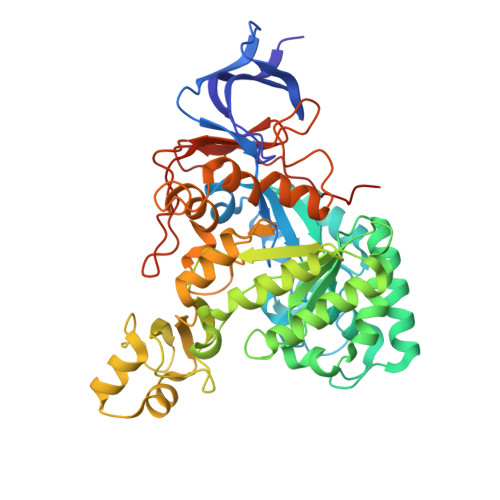
N-Acyl-D-amino acid deacylases (EC 3.5.1.81, also known as D-acylases) have been studied for decades for their utility in the kinetic resolution of N-acetyl-D,L-amino acids (NAAs) due to a marked stereospecificity. In conjunction with an N-succinyl-amino acid racemase (NSAR), they impulse the dynamic kinetic resolution (DKR) of different NAAs until the corresponding enantiomerically pure D-amino acids. Besides the clear interest in this enzyme cascade, the application of D-acylase/NSAR tandems has been only briefly described outside the industrial field. In this work, we revisit D-acylases for the DKR of NAAs, reporting the characterisation of two new recombinant D-acylases belonging to Bordetella petrii and Klebsiella pneumoniae. The enzymes were successfully coupled with the recombinant NSAR from Geobacillus stearothermophilus for the biosynthesis of D-methionine or D-aminobutyric acid. We also carried out the structural characterisation of the D-acylase from Klebsiella pneumoniae (KleDacyl), providing the second experimental 3-D structure of a member of this family of enzymes. The structural model shows a highly dynamic character of this amidohydrolase superfamily member, supplying a snapshot of an open conformation of the enzyme most likely preceding substrate entrance into the catalytic cleft. Our results confirm for the first time the importance of an α/β mobile domain in the substrate specificity of D-acylases (region 282-341 in KleDacyl), opening up new strategies for structural-based protein engineering strategies.
- Department of Biochemistry and Molecular Biology III and Immunology, University of Granada, Granada, Spain.
Organizational Affiliation: