Kinetic and structural studies of gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis.
Ferraroni, M., Angeli, A., De Luca, V., Capasso, C., Supuran, C.T.(2024) J Struct Biol 217: 108154-108154
- PubMed: 39647519
- DOI: https://doi.org/10.1016/j.jsb.2024.108154
- Primary Citation of Related Structures:
9GVA - PubMed Abstract:
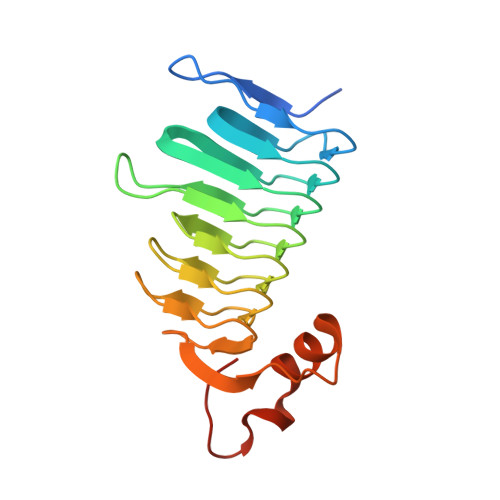
Porphyromonas gingivalis, a key pathogen in periodontal, plays a critical role in systemic pathologiesdiseases by evading host defence mechanisms and invading periodontal tissues. Targeting its virulence mechanisms and overcoming drug resistance are essential steps toward effective therapeutic development. In this study, we focused on the Carbonic Anhydrase (CA, EC: 4.2.1.1) encoded by P. gingivalis as a potential drug target. We determined the crystal structure of PgiCA γ at a resolution of 2.4 Å and conducted kinetic characterization. The structure revealed that active PgiCA γ forms a trimer, with each monomer comprising a left-handed β-helix capped by a C-terminal α-helix and coordinated to a catalytic zinc ion through three histidine residues. Interestingly, one monomer displayed an atypical α-helix conformation, likely due to close interactions with neighbouring trimers within the crystal lattice (a probable crystallographic artefact). These findings provide new insights into the structural and functional properties of PgiCA γ, emphasizing its potential as a target for the development of novel anti-virulence therapies against P. gingivalis.
- Department of Chemistry "Ugo Schiff", University of Florence, Via Della Lastruccia 3-13, 50019, Sesto Fiorentino, Italy.
Organizational Affiliation: