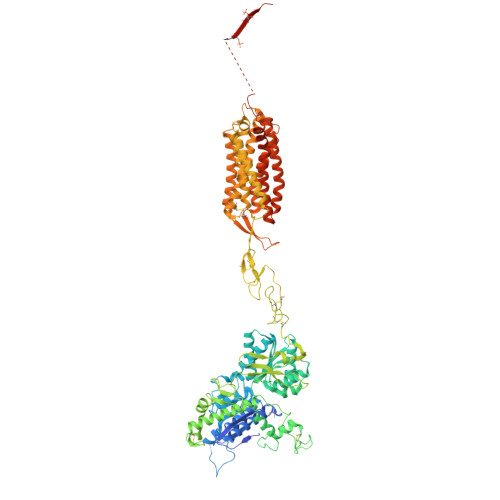
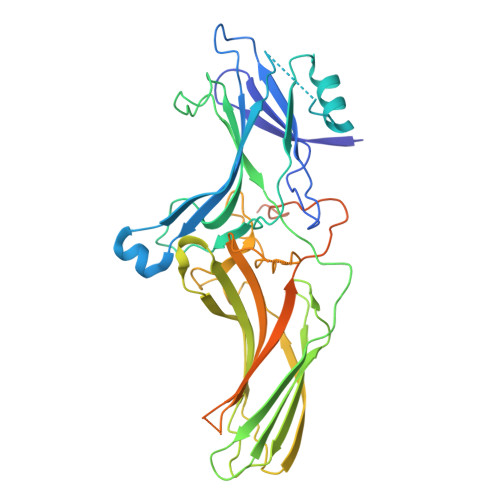
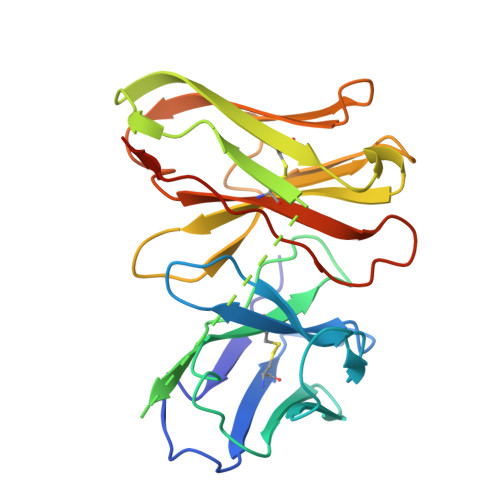
Molecular basis of beta-arrestin coupling to the metabotropic glutamate receptor mGlu3.
Wen, T., Du, M., Lu, Y., Jia, N., Lu, X., Liu, N., Chang, S., Zhang, X., Shen, Y., Yang, X.(2025) Nat Chem Biol 21: 1262-1269
- PubMed: 40050438
- DOI: https://doi.org/10.1038/s41589-025-01858-8
- Primary Citation of Related Structures:
9II2, 9II3 - PubMed Abstract:
β-Arrestins (βarrs) mediate the desensitization and internalization of activated G-protein-coupled receptors (GPCRs). The molecular mechanism by which dimeric family C GPCR members recruit arrestins remains elusive. Here we report two structures of metabotropic glutamate receptor subtype 3 (mGlu3) coupled to βarr1, with stoichiometries of 2:1 and 2:2. The L-glutamate-bound mGlu3 dimer adopts an inactive state, with both Venus flytrap domains closed, engaging βarr1 either asymmetrically or symmetrically. The transmembrane domain of the mGlu3 protomer interacts with βarr1 through a binding pocket formed by three intracellular loops and an ordered C-terminal region. Three phosphorylation sites (pS857, pS859 and pT860) on the C-terminal tail of mGlu3 engage the N domain of βarr1. βarr1 stabilizes mGlu3 in an inactive conformation, characterized by a TM3/TM4-TM3/TM4 dimeric interface, previously observed in the negative allosteric modulator-bound structure of mGlu3. Our findings provide important insights into βarr-mediated inactivation of family C GPCRs.
- State Key Laboratory of Medicinal Chemical Biology and Frontiers Science Center for Cell Responses, College of Life Sciences, Nankai University, Tianjin, China.
Organizational Affiliation: