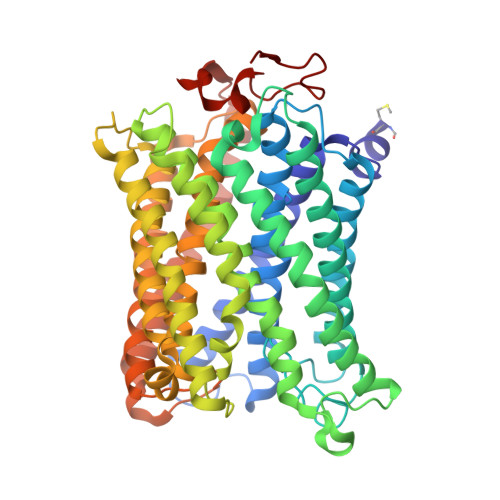
The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
Muramoto, K., Ide, T., Shinzawa-Itoh, K.(2025) J Biological Chem 301: 110395-110395
- PubMed: 40543594
- DOI: https://doi.org/10.1016/j.jbc.2025.110395
- Primary Citation of Related Structures:
9IKF, 9IKG, 9IKH, 9IKI, 9KUK, 9KUL, 9KUM, 9M56 - PubMed Abstract:
Cytochrome c oxidase (CcO) catalyzes oxygen (O 2 ) reduction at the heme a 3 -Cu B site in the transmembrane region of the enzyme. It has been proposed that the hydrophobic channel that connects the transmembrane surface of subunit III through subunit I to the heme a 3 -Cu B site is the O 2 transfer pathway. Gas molecules other than O 2 , including carbon dioxide (CO 2 ) generated in the TCA cycle, should also enter the hydrophobic channel, but it is not clear how these molecules are expelled from CcO. We analyzed the crystal structures of CO 2 -, nitrous oxide (N 2 O)-, and Xe-bound bovine CcO in the oxidized and reduced states at resolutions of 1.75-1.85 Å. Binding of Xe in the channel of subunit I near the interface with subunit III supported the proposed O 2 transfer pathway. CO 2 , N 2 O, and another Xe were all bound to a common site near the branching point of another hydrophobic channel that branched from the O 2 transport channel. Additional Xe atoms were bound in the second channel leading up to the molecular surface on the intermembrane space side, suggesting that under physiological conditions, CO 2 that has entered the O 2 pathway could be passively expelled through this channel. This channel consists of subunits I and nuclear DNA-coded subunit VIIc, suggesting that the addition of subunit VIIc in the process of molecular evolution of mitochondrial CcO has made the CO 2 exhaust pathway.
Organizational Affiliation:
Graduate School of Science, 3-2-1 Kouto, Kamigohri, Akoh, Hyogo, Japan. Electronic address: muramoto@sci.u-hyogo.ac.jp.