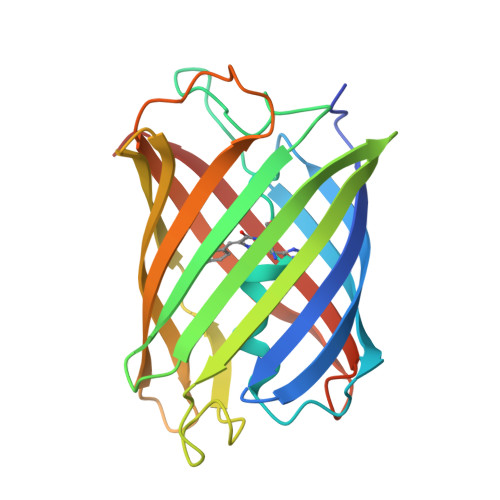
Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Zheng, S., Shi, X., Lin, J., Yang, Y., Xin, Y., Bai, X., Zhu, H., Chen, H., Wu, J., Zheng, X., Lin, L., Huang, Z., Yang, S., Hu, F., Liu, W.(2025) Acta Crystallogr D Struct Biol 81: 181-195
- PubMed: 40094266
- DOI: https://doi.org/10.1107/S2059798325002141
- Primary Citation of Related Structures:
9J0Q, 9J0R, 9J11 - PubMed Abstract:
Green-to-red photoconvertible fluorescent proteins (PCFPs) serve as key players in single-molecule localization super-resolution imaging. As an early engineered variant, mEos3.2 has limited applications, mostly due to its slow maturation rate. The recent advent of a novel variant, pcStar, obtained by the simple mutation of only three amino acids (D28E/L93M/N166G) in mEos3.2, exhibits significantly accelerated maturation and enhanced fluorescent brightness. This improvement represents an important advance in the field of biofluorescence by enabling early detection with reliable signals, essential for labelling dynamic biological processes. However, the mechanism underlying the significant improvement in fluorescent performance from mEos3.2 to pcStar remains elusive, preventing the rational design of more robust variants through mutagenesis. In this study, we determined the crystal structures of mEos3.2 and pcStar in their green states at atomic resolution and performed molecular-dynamics simulations to reveal significant divergences between the two proteins. Our structural and computational analyses revealed crucial features that are distinctively present in pcStar, including the presence of an extra solvent molecule, high conformational stability and enhanced interactions of the chromophore with its surroundings, tighter tertiary-structure packing and dynamic central-helical deformation. Resulting from the triple mutations, all of these structural features are likely to establish a mechanistic link to the greatly improved fluorescent performance of pcStar. The data described here not only provide a good example illustrating how distant amino-acid substitutions can affect the structure and bioactivity of a protein, but also give rise to strategic considerations for the future engineering of more widely applicable PCFPs.
- Key Laboratory of Gastrointestinal Cancer, Ministry of Education, School of Basic Medical Sciences, Fujian Medical University, Fuzhou, People's Republic of China.
Organizational Affiliation: