Engineering of ATP synthase for enhancement of proton-to-ATP ratio.
Ueno, H., Yasuda, K., Hamaguchi-Suzuki, N., Marui, R., Adachi, N., Senda, T., Murata, T., Noji, H.(2025) Nat Commun 16: 5410-5410
- PubMed: 40610443
- DOI: https://doi.org/10.1038/s41467-025-61227-w
- Primary Citation of Related Structures:
9JC1, 9JC2 - PubMed Abstract:
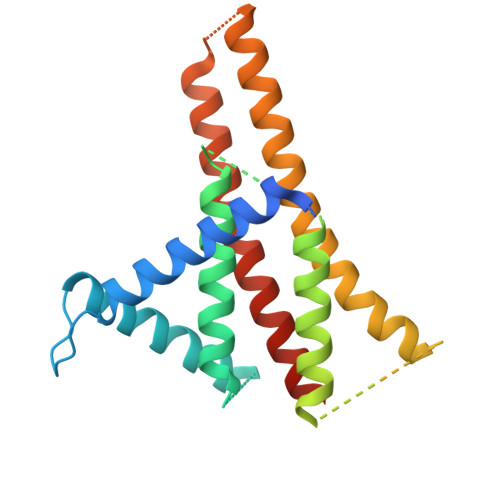
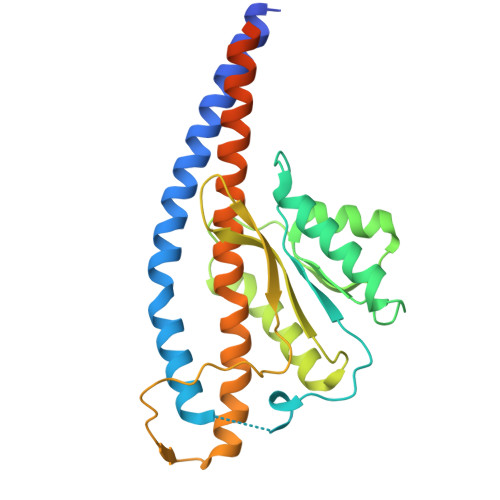
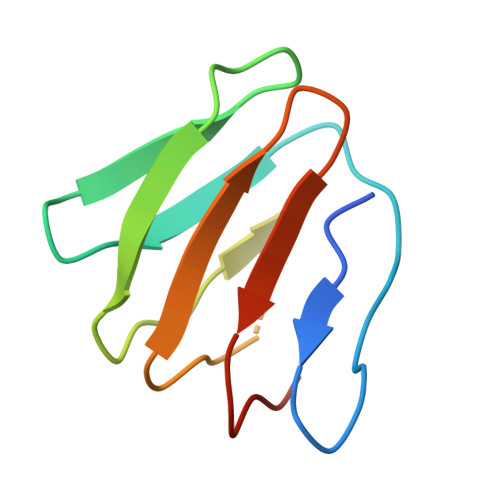
F o F 1 -ATP synthase (F o F 1 ) interconverts the energy of the proton motive force (pmf) and that of ATP through the mechanical rotation. The H + /ATP ratio, one of the most crucial parameters in bioenergetics, varies among species due to differences in the number of H + -binding c-subunits, resulting in H + /ATP ratios ranging from 2.7 to 5. In this study, we seek to significantly enhance the H + /ATP ratio by employing an alternative approach that differs from that of nature. We engineer F o F 1 to form multiple peripheral stalks, each bound to a proton-conducting a-subunit. The engineered F o F 1 exhibits an H + /ATP ratio of 5.8, surpassing the highest ratios found in naturally occurring F o F 1 s, enabling ATP synthesis under low pmf conditions where wild-type enzymes cannot synthesize ATP. Structural analysis reveals that the engineered F o F 1 forms up to three peripheral stalks and a-subunits. This study not only provides valuable insights into the H + -transport mechanism of F o F 1 but also opens up possibilities for engineering the foundation of cellular bioenergetics.
Organizational Affiliation:
Department of Applied Chemistry, Graduate School of Engineering, The University of Tokyo, Tokyo, Japan. hueno@g.ecc.u-tokyo.ac.jp.