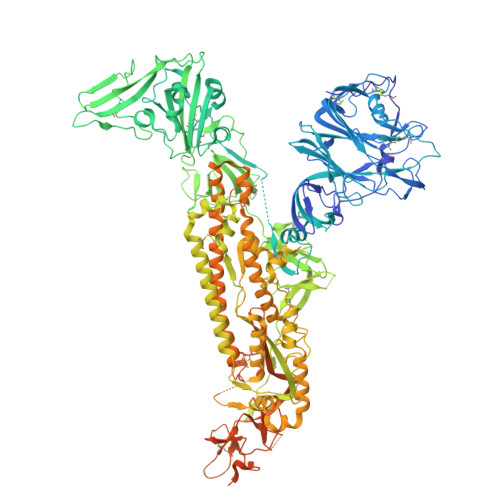
Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.
Yuan, H., Wang, J., Ma, Y., Li, Z., Gao, X., Habib, G., Liu, B., Chen, J., He, J., Zhou, P., Shi, Z.L., Chen, X., Xiong, X.(2025) Sci Adv 11: eadv7296-eadv7296
- PubMed: 40644548
- DOI: https://doi.org/10.1126/sciadv.adv7296
- Primary Citation of Related Structures:
9JMF, 9JMG, 9JMH, 9JMI, 9JMJ, 9JMM, 9JMN, 9JMO, 9JMP - PubMed Abstract:
Merbecoviruses from bats, pangolins, and hedgehogs pose significant zoonotic threats, with a limited understanding of receptor binding by their spike (S) proteins. Here, we report cryo-EM structures of GD-BatCoV (BtCoV-422) and SE-PangolinCoV (MjHKU4r-CoV-1) RBDs in complex with human DPP4 (hDPP4). These structures exhibit a substantial offset in their hDPP4 interaction interfaces, revealing a conserved hydrophobic cluster as a convergent signature of DPP4 binding within the MERS-HKU4 clade of merbecoviruses. Structure-guided mutagenesis demonstrates that favorable interactions are distributed across multiple receptor binding motif (RBM) regions, working synergistically to confer high-affinity hDPP4 binding. Swapping of the merbecovirus RBM regions indicate limited plasticity and interchangeability among these regions. In addition, we report cryo-EM structures of six merbecovirus S-trimers. Structure-based phylogenetics suggests that hDPP4-binding merbecoviruses undergo convergent evolution, while ACE2-binding merbecoviruses exhibit diversification in their binding mechanisms. These findings offer critical insights into merbecovirus receptor utilization, providing a structural understanding for future surveillance.
- State Key Laboratory of Respiratory Disease, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China.
Organizational Affiliation: