Real-time capture of sigma N transcription initiation intermediates reveals mechanism of ATPase-driven activation by limited unfolding.
Mueller, A.U., Molina, N., Nixon, B.T., Darst, S.A.(2025) Nat Commun 16: 7138-7138
- PubMed: 40759887
- DOI: https://doi.org/10.1038/s41467-025-61837-4
- Primary Citation of Related Structures:
9MSE, 9MSF, 9MSG, 9MSH, 9MSJ - PubMed Abstract:
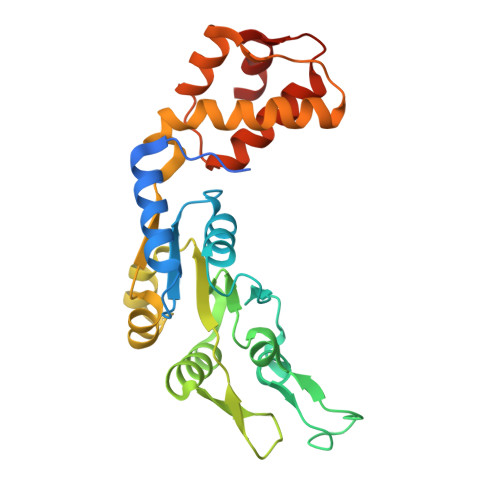
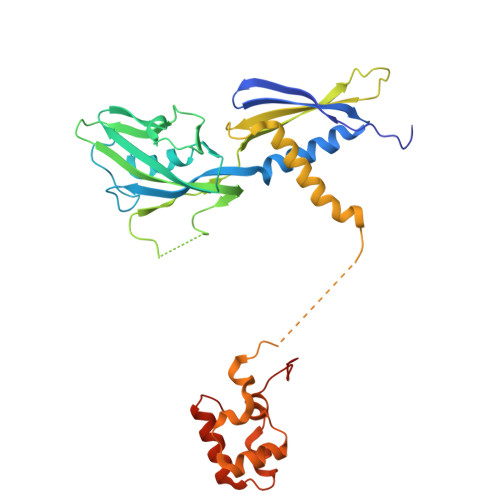
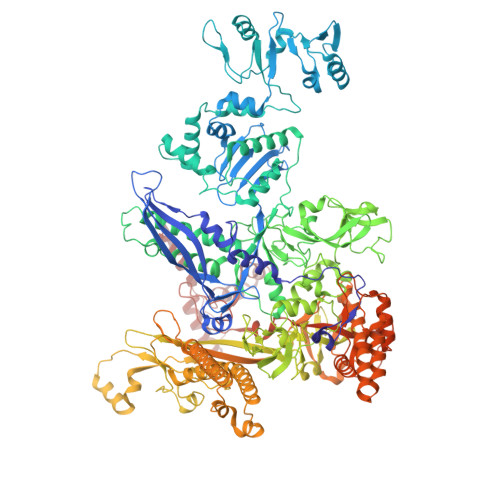
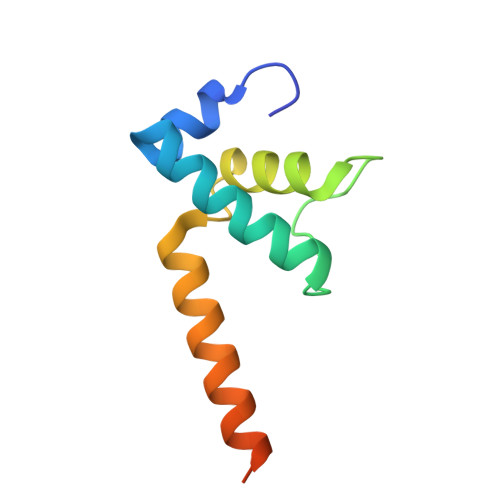
Bacterial σ factors bind RNA polymerase (E) to form holoenzyme (Eσ), conferring promoter specificity to E and playing a key role in transcription bubble formation. σ N is unique among σ factors in its structure and functional mechanism, requiring activation by specialized AAA+ ATPases. Eσ N forms an inactive promoter complex where the N-terminal σ N region I (σ N -RI) threads through a small DNA bubble. On the opposite side of the DNA, the ATPase engages σ N -RI within the pore of its hexameric ring. Here, we perform kinetics-guided structural analysis of de novo formed Eσ N initiation complexes and engineer a biochemical assay to measure ATPase-mediated σ N -RI translocation during promoter melting. We show that the ATPase exerts mechanical action to translocate about 30 residues of σ N -RI through the DNA bubble, disrupting inhibitory structures of σ N to allow full transcription bubble formation. A local charge switch of σ N -RI from positive to negative may help facilitate disengagement of the otherwise processive ATPase, allowing subsequent σ N disentanglement from the DNA bubble.
- Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY, USA.
Organizational Affiliation: