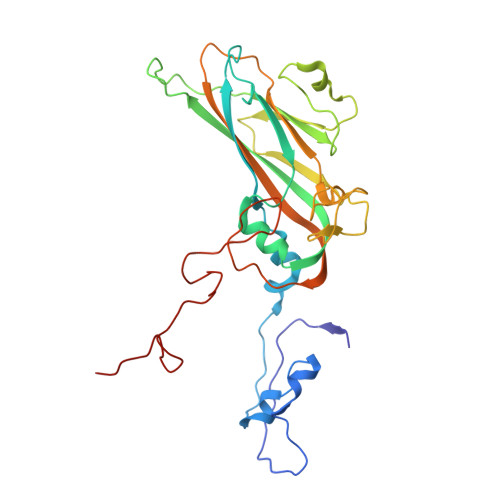
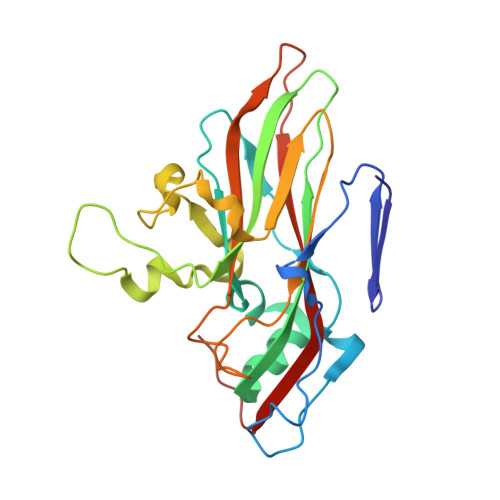
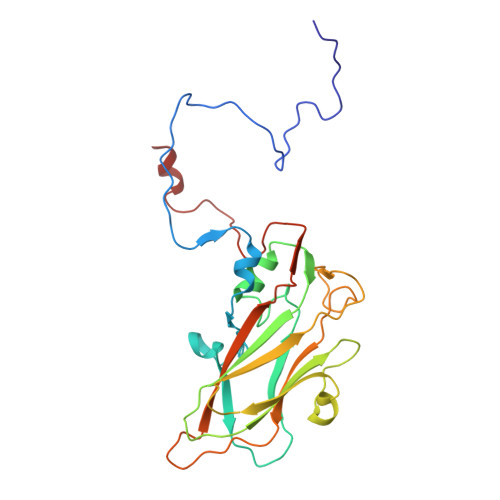
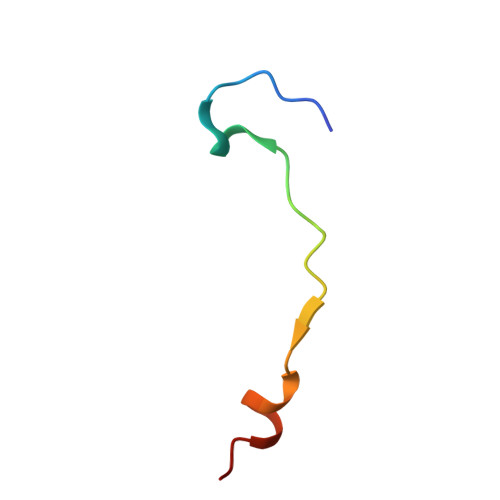
MFSD6 is an entry receptor for enterovirus D68.
Varanese, L., Xu, L., Peters, C.E., Pintilie, G., Roberts, D.S., Raj, S., Liu, M., Ooi, Y.S., Diep, J., Qiao, W., Richards, C.M., Callaway, J., Bertozzi, C.R., Jabs, S., de Vries, E., van Kuppeveld, F.J.M., Nagamine, C.M., Chiu, W., Carette, J.E.(2025) Nature 641: 1268-1275
- PubMed: 40132641
- DOI: https://doi.org/10.1038/s41586-025-08908-0
- Primary Citation of Related Structures:
9MWZ, 9MXC - PubMed Abstract:
With the near eradication of poliovirus due to global vaccination campaigns, attention has shifted to other enteroviruses that can cause polio-like paralysis syndrome (now termed acute flaccid myelitis (AFM)) 1-3 . In particular, enterovirus D68 (EV-D68) is believed to be the main driver of epidemic outbreaks of AFM in recent years 4 , yet not much is known about EV-D68 host interactions. EV-D68 is a respiratory virus 5 but, in rare cases, can spread to the central nervous system to cause severe neuropathogenesis. Here, we used genome-scale CRISPR screens to identify the poorly characterized multipass membrane transporter MFSD6 as a host entry factor for EV-D68. Knockout of MFSD6 expression abrogated EV-D68 infection in cell lines and primary cells corresponding to respiratory and neural cells. MFSD6 localized to the plasma membrane and was required for viral entry into host cells. MFSD6 bound directly to EV-D68 particles via its third extracellular loop (L3). We determined the cryo-EM structure of EV-D68 in complex with L3 at 2.1 Å resolution, revealing the interaction interface. A decoy receptor, engineered by fusing MFSD6(L3) to Fc, blocked EV-D68 infection of human primary lung epithelial cells, and provided near complete protection in a lethal mouse model of EV-D68 infection. Collectively, our results reveal MFSD6 as an entry receptor for EV-D68, and support targeting MFSD6 as a potential mechanism to combat infections by this emerging pathogen with pandemic potential.
- Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, CA, USA.
Organizational Affiliation: