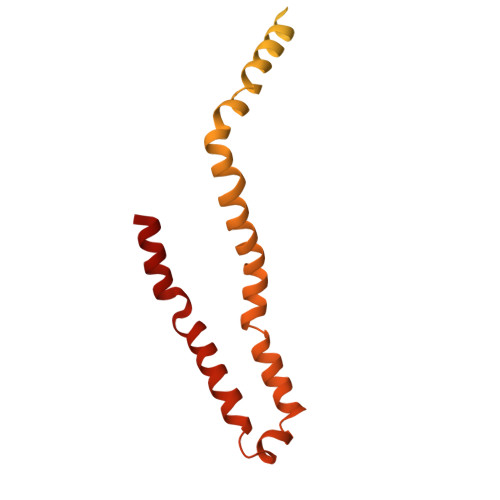
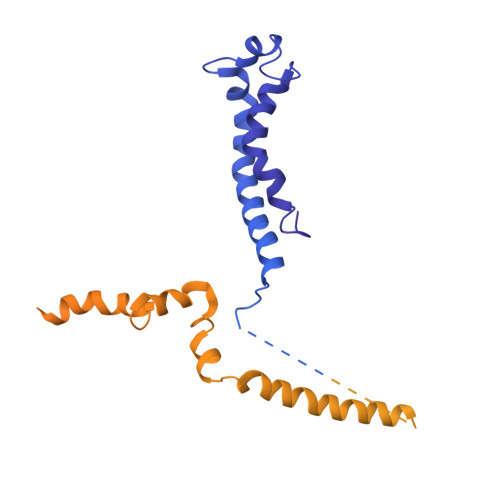
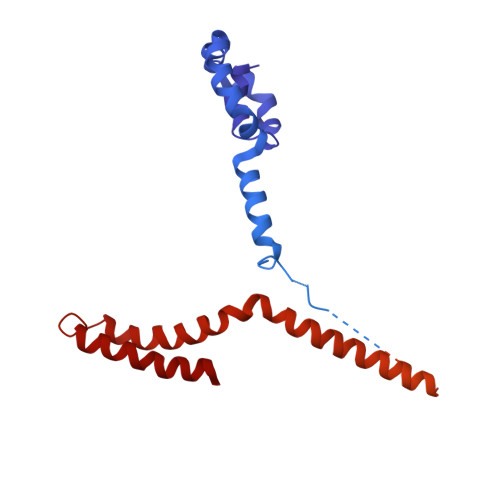
Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.
Ashaduzzaman, M., Taheri, A., Lee, Y.J., Tang, Y., Guo, F., Fried, S.D., Liu, B., Al-Bassam, J.(2025) bioRxiv
- PubMed: 40034650
- DOI: https://doi.org/10.1101/2025.02.25.640204
- Primary Citation of Related Structures:
9NA8, 9NA9, 9NBA, 9NBB, 9NBD, 9NBI - PubMed Abstract:
Microtubule (MT) branch nucleation is fundamental for building parallel MT networks in eukaryotic cells. In plants and metazoans, MT branch nucleation requires Augmin and NEDD1 proteins which bind along MTs and then recruit and activate the gamma-tubulin ring complex (γ-TuRC). Augmin is a fork-shaped assembly composed of eight coiled-coil subunits, while NEDD1 is a WD40 β-propellor protein that bridges across MTs, Augmin, and γ-TuRC during MT branch nucleation. Here, we reconstitute hetero-tetrameric and hetero-octameric Arabidopsis thaliana Augmin assemblies, resolve their subunit interactions using crosslinking mass spectrometry and determine 3.7 to 7.3-Å cryo-EM structures for the V-junction and extended regions of Augmin. These structures allowed us to generate a complete de novo plant Augmin model that reveals the long-range multi coiled-coil interfaces that stabilize its 40-nm hetero-octameric fork-shaped organization. We discovered the dual calponin homology (CH) domain forming its MT binding site at the end of its V-junction undertake open and closed conformations. We determined a 12-Å dimeric Augmin cryo-EM structure revealing Augmin undergoes anti-parallel dimerization through two conserved surfaces along Augmin's extended region. We reconstituted the NEDD1 WD40 β-propellor with Augmin revealing it directly binds on top its V-junction and enhances Augmin dimerization. Our studies suggest that cooperativity between the Augmin dual CH domains and NEDD1 WD40 binding site may regulate Augmin V-junction dual binding to MT lattices. This unique V-shaped dual binding and organization anchors Augmins along MTs generating a platform to recruit γ-TuRC and activate branched MT nucleation.
- Department of Molecular Cellular Biology, University of California, Davis, CA, USA.
Organizational Affiliation: