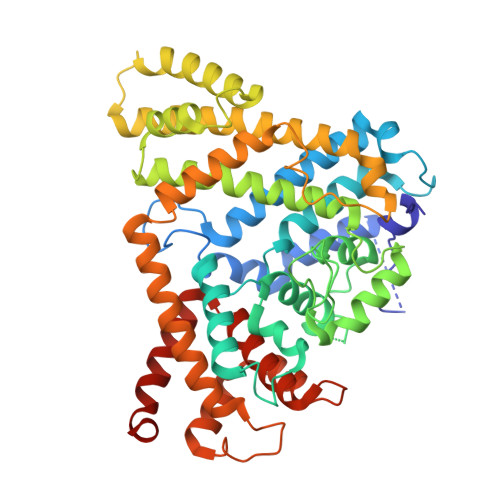
Activating and inhibiting nucleotide signals coordinate bacterial anti-phage defense.
Yamaguchi, S., Fernandez, S.G., Wassarman, D.R., Luders, M., Schwede, F., Kranzusch, P.J.(2025) bioRxiv
- PubMed: 40672243
- DOI: https://doi.org/10.1101/2025.07.09.663793
- Primary Citation of Related Structures:
9P8S, 9P8T, 9P8U, 9P8V, 9P8W - PubMed Abstract:
The cellular nucleotide pool is a major focal point of the host immune response to viral infection. Immune effector proteins that disrupt the nucleotide pool allow animal and bacterial cells to broadly restrict diverse viruses, but reduced nucleotide availability induces cellular toxicity and can limit host fitness(Ahmad et al., 1998; Goldstone et al., 2011; Hsueh et al., 2022; Itsko & Schaaper, 2014; Tal et al., 2022). Here we discover a bacterial anti-phage defense system named Clover that overcomes this tradeoff by encoding a deoxynucleoside triphosphohydrolase enzyme (CloA) that dynamically responds to both an activating phage cue and an inhibitory nucleotide immune signal produced by a partnering regulatory enzyme (CloB). Analysis of Clover phage restriction in cells and reconstitution of enzymatic function in vitro demonstrate that CloA is a dGTPase that responds to viral enzymes that increase cellular levels of dTTP. To restrain CloA activation in the absence of infection, we show that CloB synthesizes a dTTP-related inhibitory nucleotide signal p3diT (5'-triphosphothymidyl-3'5'-thymidine) that binds to CloA and suppresses activation. Cryo-EM structures of CloA in activated and suppressed states reveal how dTTP and p3diT control distinct allosteric sites and regulate effector function. Our results define how nucleotide signals coordinate both activation and inhibition of antiviral immunity and explain how cells balance defense and immune-mediated toxicity.
- Department of Microbiology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: