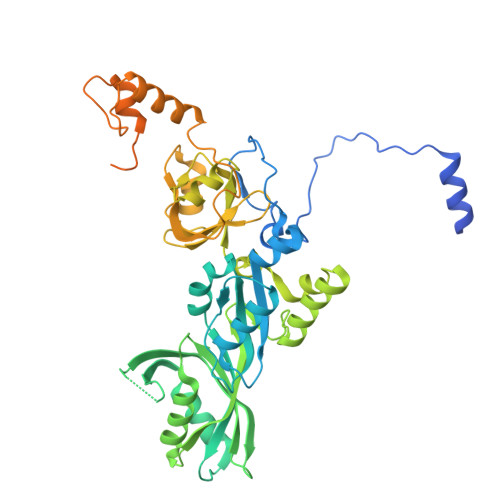
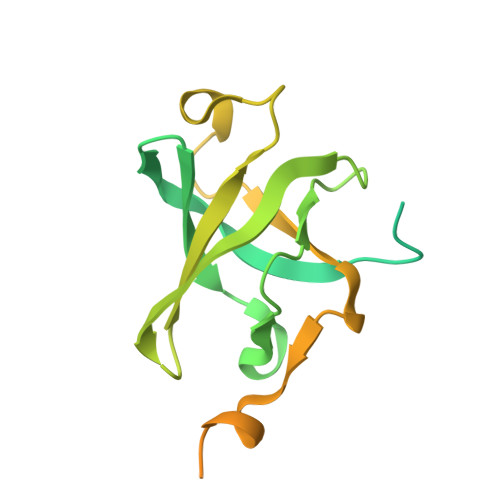
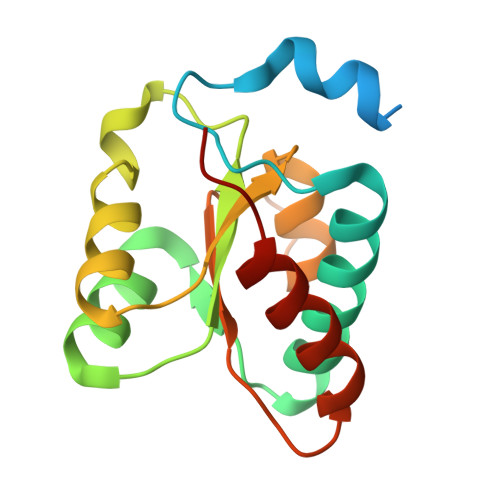
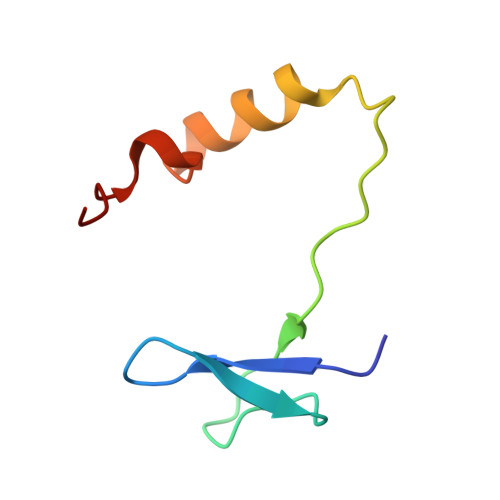
Cryo-EM structure of human telomerase dimer reveals H/ACA RNP-mediated dimerization.
Balch, S., Sekne, Z., Franco-Echevarria, E., Ludzia, P., Kretsch, R.C., Sun, W., Yu, H., Ghanim, G.E., Thorkelsson, S., Ding, Y., Das, R., Nguyen, T.H.D.(2025) Science 389: eadr5817-eadr5817
- PubMed: 40638752
- DOI: https://doi.org/10.1126/science.adr5817
- Primary Citation of Related Structures:
9QAX, 9QAY, 9QAZ, 9QB2, 9QB3 - PubMed Abstract:
Telomerase ribonucleoprotein (RNP) synthesizes telomeric repeats at chromosome ends using a telomerase reverse transcriptase (TERT) and a telomerase RNA (hTR in humans). Previous structural work showed that human telomerase is typically monomeric, containing a single copy of TERT and hTR. Evidence for dimeric complexes exists, although the composition, high-resolution structure, and function remain elusive. Here, we report the cryo-electron microscopy (cryo-EM) structure of a human telomerase dimer bound to telomeric DNA. The structure reveals a 26-subunit assembly and a dimerization interface mediated by the Hinge and ACA box (H/ACA) RNP of telomerase. Premature aging disease mutations map to this interface. Disrupting dimer formation affects RNP assembly, bulk telomerase activity, and telomere maintenance in cells. Our findings address a long-standing enigma surrounding the telomerase dimer and suggest a role for the dimer in telomerase assembly.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: