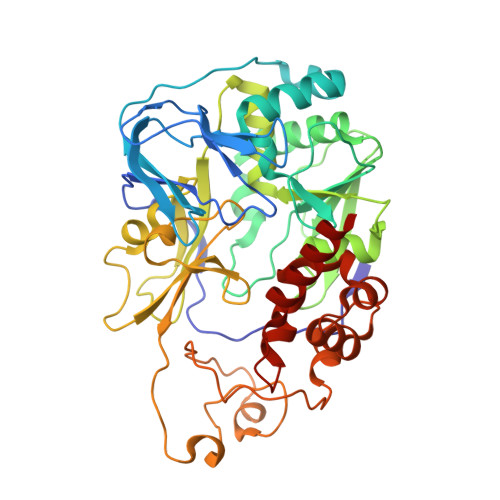
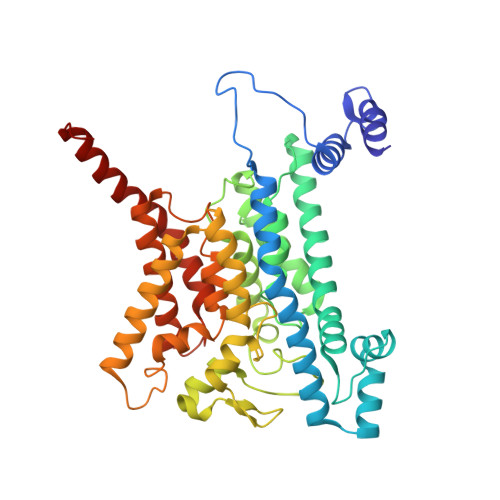
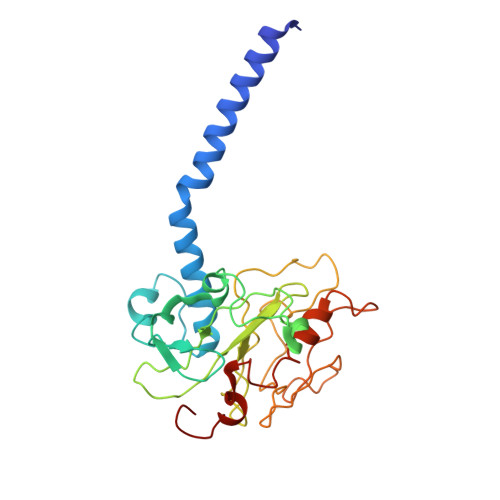
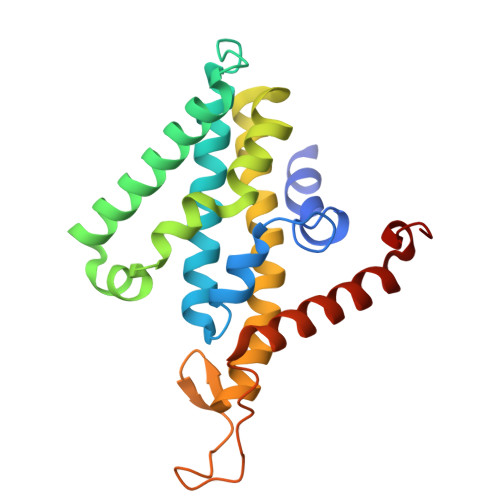
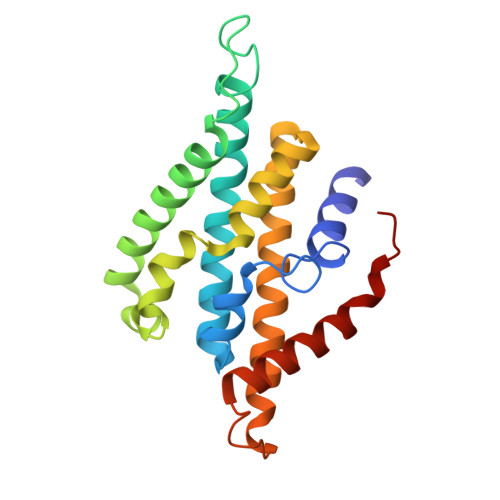
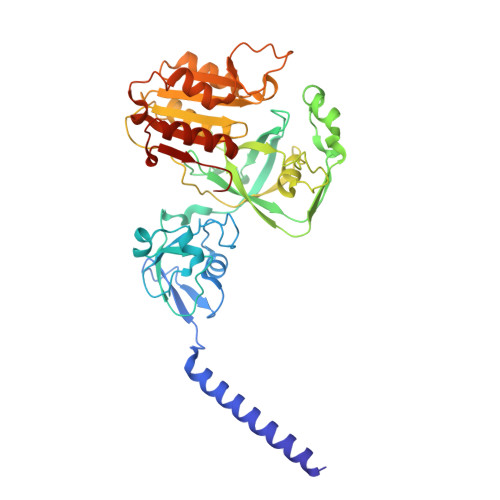
The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Ishikawa-Fukuda, M., Seki, T., Kishikawa, J.I., Takahiro, M., Okazaki, K.I., Kato, T., Barquera, B., Miyoshi, H., Murai, M.(2025) bioRxiv
- PubMed: 40501732
- DOI: https://doi.org/10.1101/2025.06.01.656757
- Primary Citation of Related Structures:
9U5G, 9UD2, 9UD3, 9UD4, 9UD5, 9UD6, 9UD8, 9UD9, 9UDA, 9UDF, 9UDG, 9UUU - PubMed Abstract:
The Na + -pumping NADH-quinone oxidoreductase (Na + -NQR) is a key respiratory enzyme in many marine and pathogenic bacteria that couples electron transfer to Na + -pumping across the membrane. Earlier X-ray and cryo-EM structures of Na + -NQR from Vibrio cholerae suggested that the subunits harboring redox cofactors undergo conformational changes during catalytic turnover. However, these proposed rearrangements have not yet been confirmed. Here, we have identified at least five distinct conformational states of Na + -NQR using: mutants that lack specific cofactors, specific inhibitors or low-sodium conditions. Molecular dynamics simulations based on these structural insights indicate that 2Fe-2S reduction in NqrD/E plays a crucial role in triggering Na + translocation by driving structural rearrangements in the NqrD/E subunits, which subsequently influence NqrC and NqrF positioning. This study provides the first structural insights into the mechanism of Na + translocation coupled to electron transfer in Na⁺-NQR.