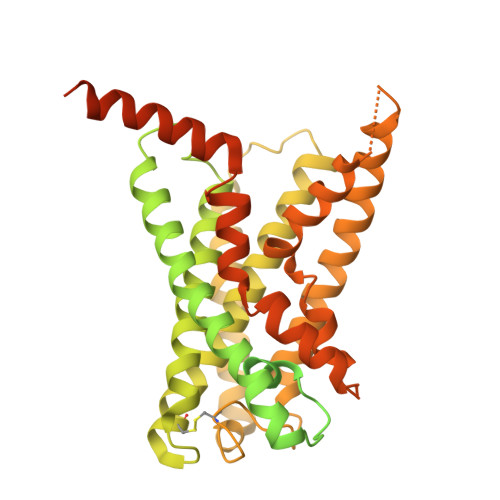
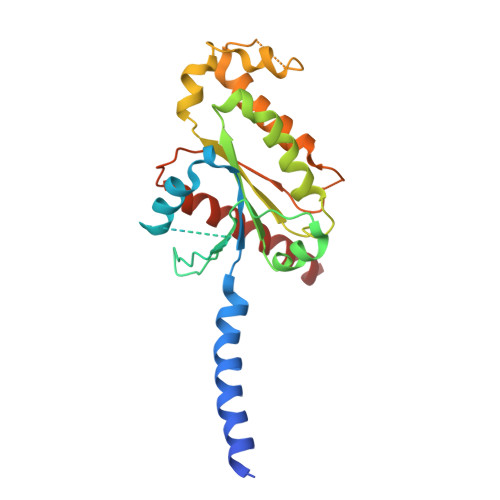
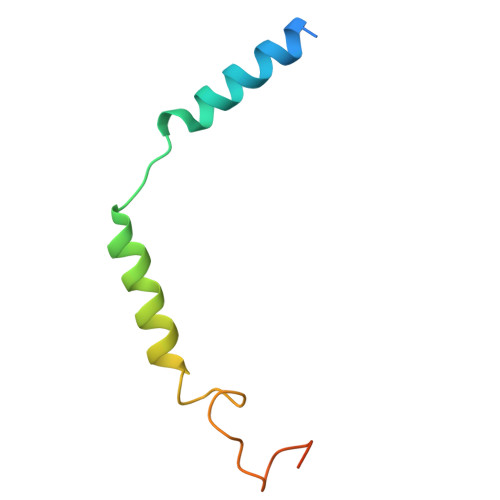
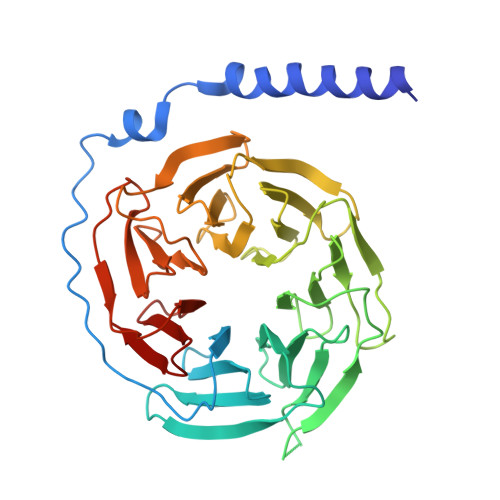
Cryo-EM structural elucidation and molecular mechanism of the GPR133-G13 signaling complex.
Pu, X., Xi, Y.T., Wang, M.X., Zhang, D., Ping, Y.Q., Xiao, P., Sun, J.P.(2025) Biochem Biophys Res Commun 777: 152165-152165
- PubMed: 40570642
- DOI: https://doi.org/10.1016/j.bbrc.2025.152165
- Primary Citation of Related Structures:
9V0U - PubMed Abstract:
GPR133 is an adhesion-class G protein-coupled receptor (GPCR) that has recently been de-orphanized. Its functions are complex and multifaceted. While GPR133 is primarily recognized for coupling with the Gs subunit to mediate elevated intracellular cAMP levels, its potential engagement with alternative signaling pathways remains poorly characterized. In our experiments, we demonstrated that GPR133 exhibits constitutive self-activation via its Stachel sequence as an adhesion GPCR, enabling activation of downstream G13 signaling. We reconstituted the GPR133-GAIN-miniGα13 complex in vitro and resolved its cryo-electron microscopy structure at a resolution of 3.51 Å. Detailed structural comparisons between the GPR133-GAIN-miniGα13 complex and the previously resolved GPR133-CTF-Gs structure highlighted both conserved and different features. These findings provide critical insights into the signal transduction mechanisms of GPR133 and lay a foundation for targeted therapeutic strategies.
- School of Pharmacy, Binzhou Medical University, 264003, Yantai, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan, 250012, China.
Organizational Affiliation: