Crystal structure of Isoform Chitin Binding Protein from Iberis umbellata L.
Saeed, A., Betzel, C., Brognaro, H., Rajaiah Prabhu, P., Alves Franca, B., Mehmood, S., Khaliq, B., Ishaq, U., Akrem, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chitin Binding Protein III | 35 | Iberis umbellata | Mutation(s): 0 | 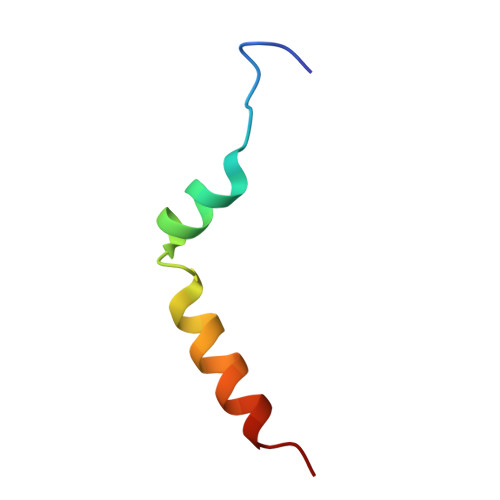 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chitin Binding Protein III | 74 | Iberis umbellata | Mutation(s): 0 | 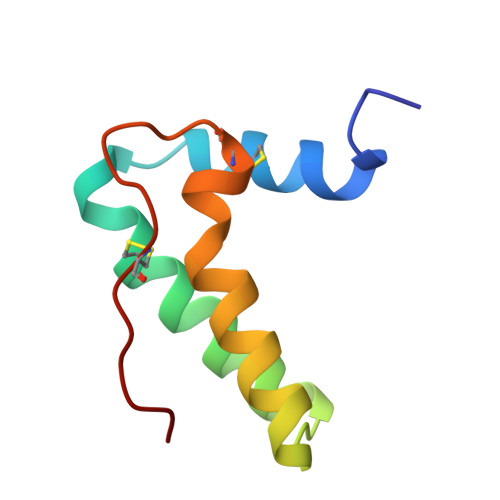 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 (Subject of Investigation/LOI) Query on SO4 | C [auth A], J [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| NO3 (Subject of Investigation/LOI) Query on NO3 | F [auth A] | NITRATE ION N O3 NHNBFGGVMKEFGY-UHFFFAOYSA-N |  | ||
| ACT (Subject of Investigation/LOI) Query on ACT | L [auth B], M [auth B] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| CL (Subject of Investigation/LOI) Query on CL | G [auth B], H [auth B], I [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA (Subject of Investigation/LOI) Query on NA | D [auth A], E [auth A], K [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.179 | α = 90 |
| b = 73.121 | β = 90 |
| c = 31.296 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| pointless | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | Pakistan | IRSIP |