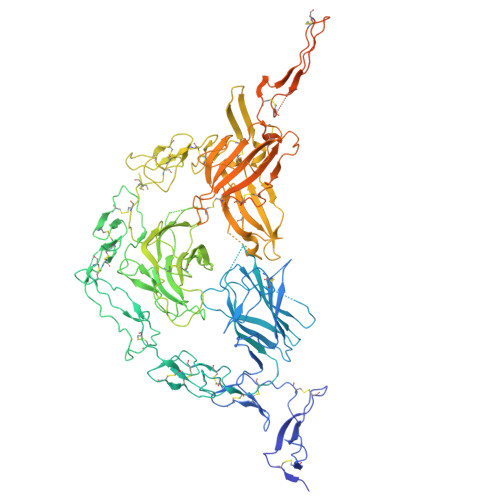
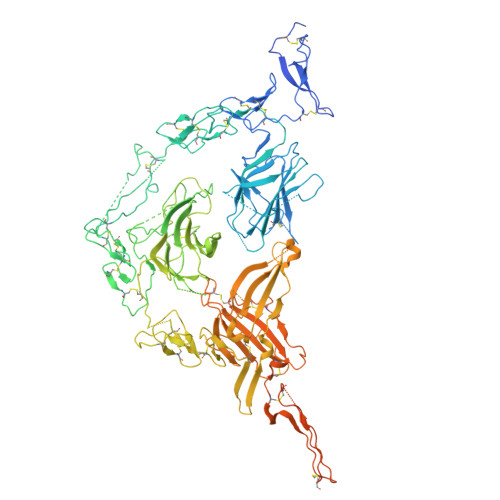
ELAPOR1 is a copper-dependent tethering factor driving proacrosomal vesicle fusion during acrosome biogenesis.
Shao, T., Ma, J., Tan, X., Shan, H., Xu, D., Zhang, K., Zheng, S., Wang, F.(2025) Proc Natl Acad Sci U S A 122: e2501302122-e2501302122
- PubMed: 40737321
- DOI: https://doi.org/10.1073/pnas.2501302122
- Primary Citation of Related Structures:
9VQL, 9VQM - PubMed Abstract:
The acrosome is a crucial organelle essential for sperm function and male fertility. During acrosome biogenesis, numerous proacrosomal vesicles (PAVs) are transported to the concave region of the nuclear membrane and fuse to form the acrosome. However, the mechanisms governing the fusion of PAVs to form the acrosome remain poorly understood. Here, we identify endosome-lysosome associated apoptosis and autophagy regulator 1 (ELAPOR1), a conserved protein, as a key factor in PAVs fusion during acrosome biogenesis. Male mice lacking Elapor1 ( Elapor1 -/- ) are infertile, exhibiting defective acrosome biogenesis and a globozoospermia-like phenotype. Using cryo-electron microscopy revealed that ELAPOR1 forms a square planar homodimer in cis, which assembles into a trans-tetramer via head-to-head homophilic interactions dependent on copper chelation. Notably, ELAPOR1 exhibits dual membrane orientation, with a predicted N in - C out topology and a noncanonical N out - C in topology in vesicles. The noncanonical N out - C in topology enables ELAPOR1 to function as a tethering factor bridging vesicles through head-to-head homophilic interactions. A mutant ELAPOR1 (ELAPOR1 4HA ) incapable of copper chelation forms cis homodimers but fails to mediate homophilic interactions in vitro, leading to defective PAVs fusion in mice, phenocopying the Elapor1 -deficient mice. Additionally, ELAPOR1 was shown to interact with soluble N-ethylmaleimide sensitive factor attachment protein receptors protein STX12. Conditional knockout of Stx12 in germ cells resulted in similar defects in acrosome biogenesis. Collectively, our findings suggest that ELAPOR1 functions as a tethering factor that regulates PAV fusion through a copper-dependent mechanism.
- Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China.
Organizational Affiliation: