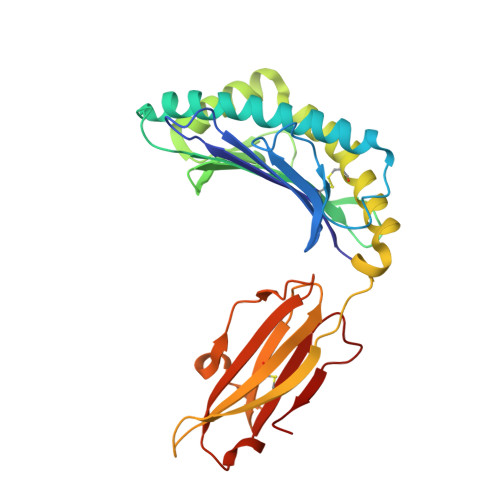
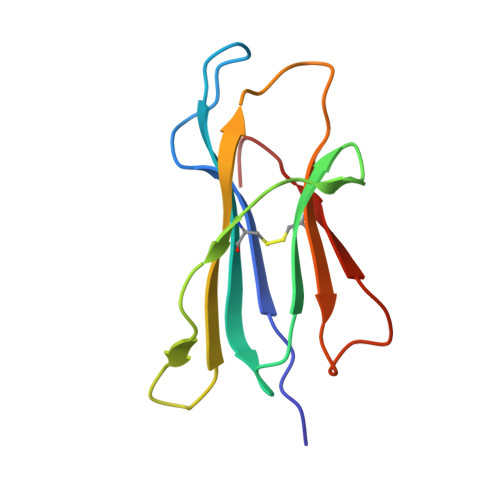
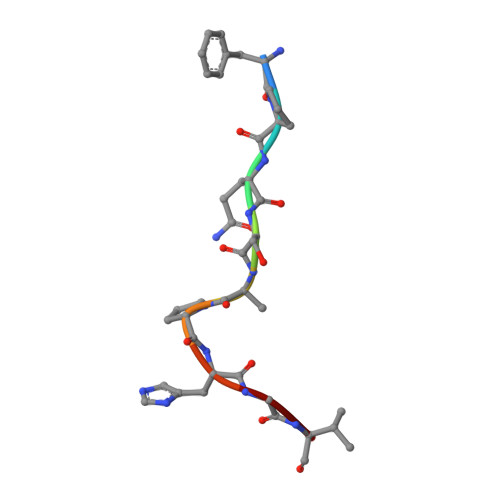
Amino acid insertion in Bat MHC-I enhances complex stability and augments peptide presentation.
Wang, S., Zheng, L., Wei, X., Qu, Z., Du, L., Wang, S., Zhang, N.(2024) Commun Biol 7: 586-586
- PubMed: 38755285
- DOI: https://doi.org/10.1038/s42003-024-06292-5
- Primary Citation of Related Structures:
8HSM, 8HSO, 8HSW, 8HT1, 8HT9 - PubMed Abstract:
Bats serve as reservoirs for numerous zoonotic viruses, yet they typically remain asymptomatic owing to their unique immune system. Of particular significance is the MHC-I in bats, which plays crucial role in anti-viral response and exhibits polymorphic amino acid (AA) insertions. This study demonstrated that both 5AA and 3AA insertions enhance the thermal stability of the bat MHC-I complex and enrich the diversity of bound peptides in terms of quantity and length distribution, by stabilizing the 3 10 helix, a region prone to conformational changes during peptide loading. However, the mismatched insertion could diminish the stability of bat pMHC-I. We proposed that a suitable insertion may help bat MHC-I adapt to high body temperatures during flight while enhancing antiviral responses. Moreover, this site-specific insertions may represent a strategy of evolutionary adaptation of MHC-I molecules to fluctuations in body temperature, as similar insertions have been found in other lower vertebrates.
- National Key Laboratory of Veterinary Public Health Security, Key Laboratory of Animal Epidemiology of the Ministry of Agriculture and Rural Affairs, College of Veterinary Medicine, China Agricultural University, Beijing, 100193, PR China.
Organizational Affiliation: