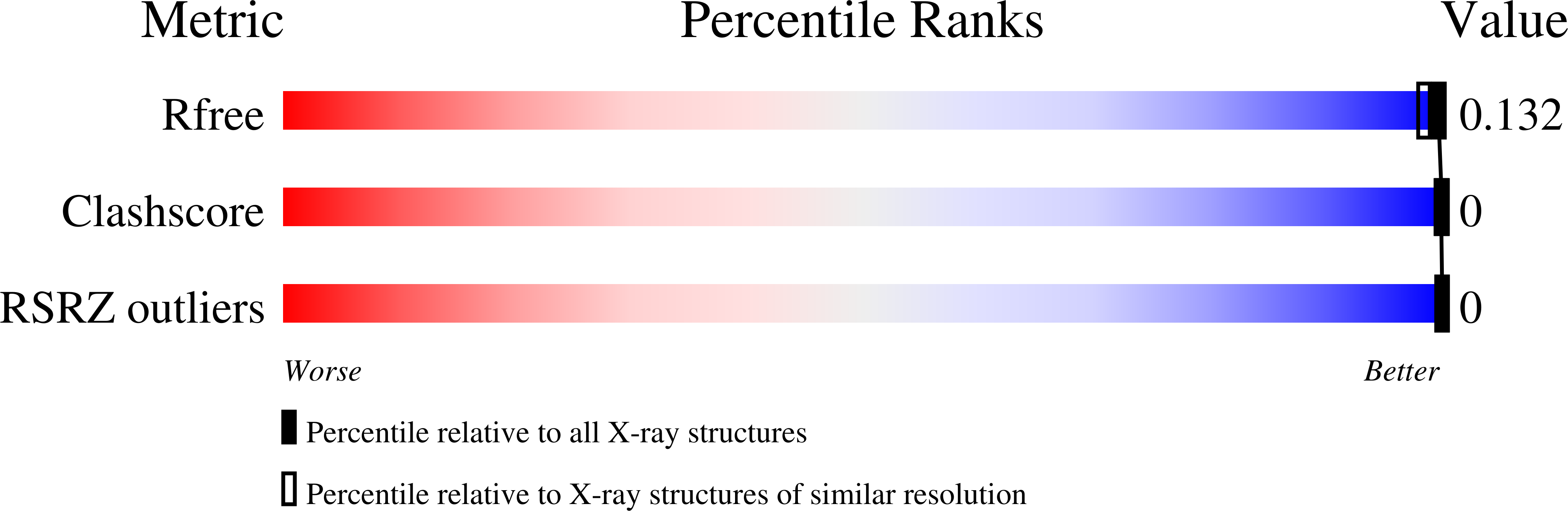Monitoring one-electron photo-oxidation of guanine in DNA crystals using ultrafast infrared spectroscopy.
Hall, J.P., Poynton, F.E., Keane, P.M., Gurung, S.P., Brazier, J.A., Cardin, D.J., Winter, G., Gunnlaugsson, T., Sazanovich, I.V., Towrie, M., Cardin, C.J., Kelly, J.M., Quinn, S.J.(2015) Nat Chem 7: 961-967
- PubMed: 26587711
- DOI: https://doi.org/10.1038/nchem.2369
- Primary Citation of Related Structures:
4R8J - PubMed Abstract:
To understand the molecular origins of diseases caused by ultraviolet and visible light, and also to develop photodynamic therapy, it is important to resolve the mechanism of photoinduced DNA damage. Damage to DNA bound to a photosensitizer molecule frequently proceeds by one-electron photo-oxidation of guanine, but the precise dynamics of this process are sensitive to the location and the orientation of the photosensitizer, which are very difficult to define in solution. To overcome this, ultrafast time-resolved infrared (TRIR) spectroscopy was performed on photoexcited ruthenium polypyridyl-DNA crystals, the atomic structure of which was determined by X-ray crystallography. By combining the X-ray and TRIR data we are able to define both the geometry of the reaction site and the rates of individual steps in a reversible photoinduced electron-transfer process. This allows us to propose an individual guanine as the reaction site and, intriguingly, reveals that the dynamics in the crystal state are quite similar to those observed in the solvent medium.
Organizational Affiliation:
Department of Chemistry, University of Reading, Whiteknights, Reading RG6 6AD, UK.