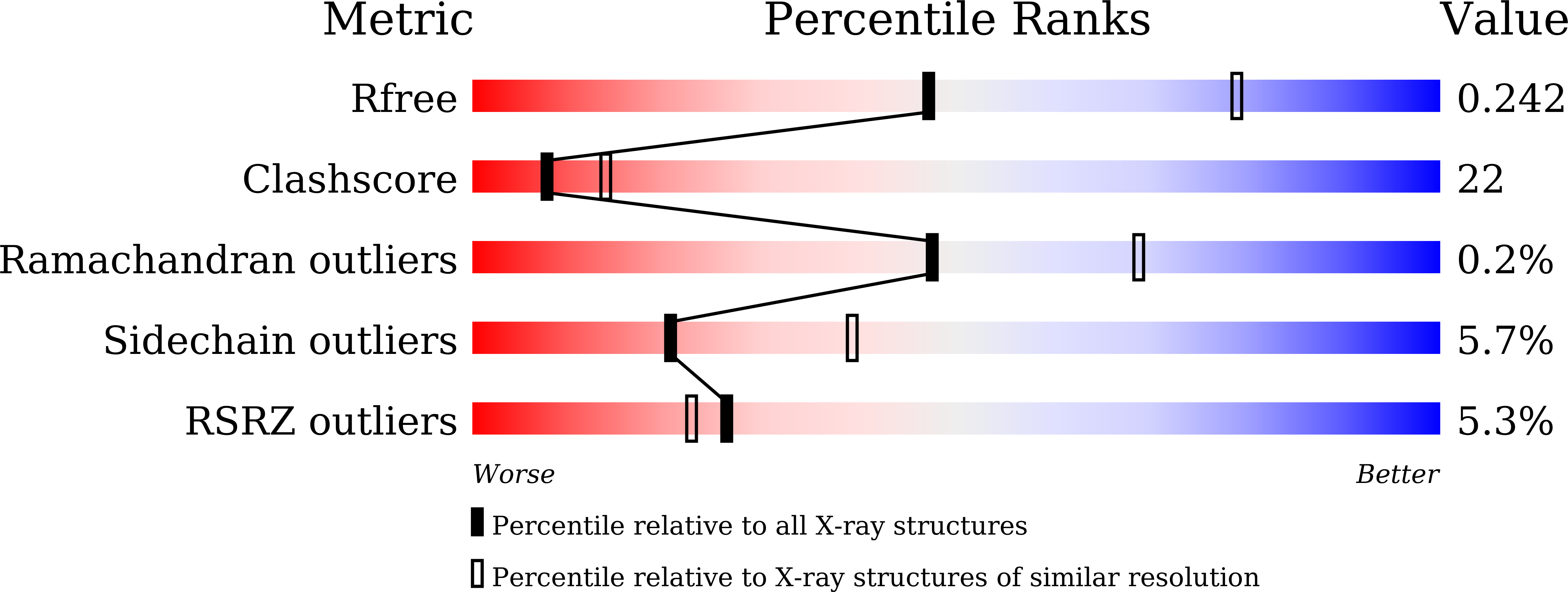
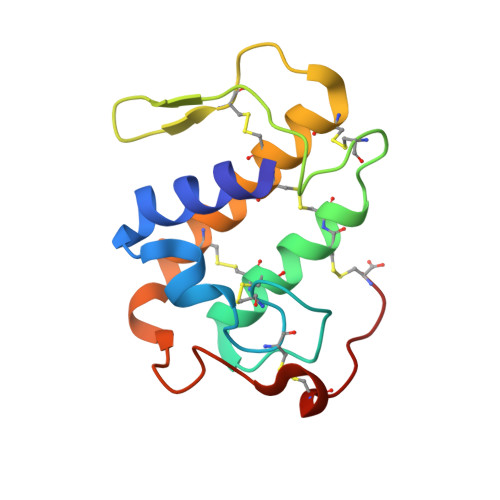
SDS-induced oligomerization of Lys49-phospholipase A2from snake venom.
Matsui, T., Kamata, S., Ishii, K., Maruno, T., Ghanem, N., Uchiyama, S., Kato, K., Suzuki, A., Oda-Ueda, N., Ogawa, T., Tanaka, Y.(2019) Sci Rep 9: 2330-2330
- PubMed: 30787342
- DOI: https://doi.org/10.1038/s41598-019-38861-8
- Primary Citation of Related Structures:
6AL3 - PubMed Abstract:
Phospholipase A 2 (PLA 2 ) is one of the representative toxic components of snake venom. PLA 2 s are categorized into several subgroups according to the amino acid at position 49, which comprises either Asp49, Lys49, Arg49 or Ser49. Previous studies suggested that the Lys49-PLA 2 assembles into an extremely stable dimer. Although the behavior on Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing or non-reducing conditions suggested the presence of intermolecular disulfide bonds, these bonds were not observed in the crystal structure of Lys49-PLA 2 . The reason for this discrepancy between the crystal structure and SDS-PAGE of Lys49-PLA 2 remains unknown. In this study, we analyzed a Lys49-PLA 2 homologue from Protobothrops flavoviridis (PflLys49-PLA 2 BPII), by biophysical analyses including X-ray crystallography, SDS-PAGE, native-mass spectrometry, and analytical ultracentrifugation. The results demonstrated that PflLys49-PLA 2 BPII spontaneously oligomerized in the presence of SDS, which is one of the strongest protein denaturants.
Organizational Affiliation:
Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577, Japan.