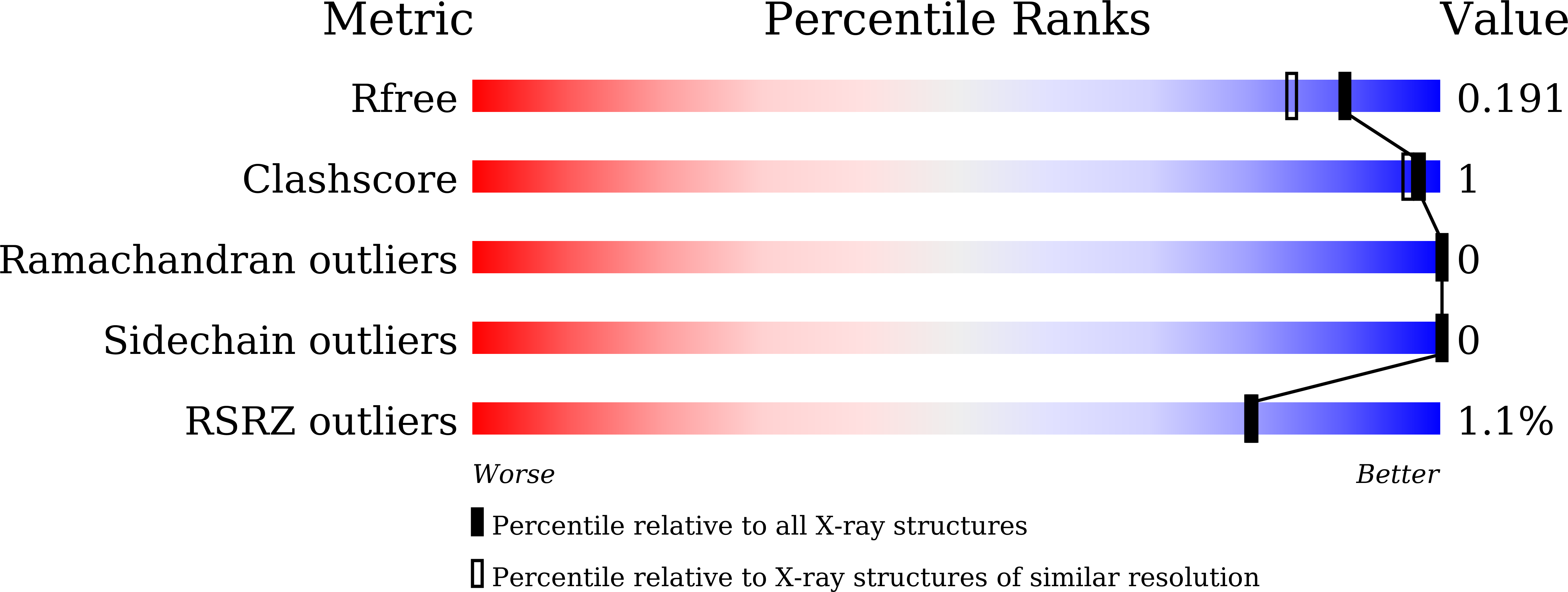
Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Zhang, C., Min, Z., Liu, X., Wang, C., Wang, Z., Shen, J., Tang, W., Zhang, X., Liu, D., Xu, X.(2020) FEBS Lett 594: 564-580
- PubMed: 31573681
- DOI: https://doi.org/10.1002/1873-3468.13630
- Primary Citation of Related Structures:
6KIK, 6KIY, 6KY6 - PubMed Abstract:
Tolrestat and epalrestat have been characterized as noncompetitive inhibitors of aldo-ketone reductase 1B1 (AKR1B1), a leading drug target for the treatment of type 2 diabetes complications. However, clinical applications are limited for most AKR1B1 inhibitors due to adverse effects of cross-inhibition with other AKRs. Here, we report an atypical competitive binding and inhibitory effect of tolrestat on the thermostable AKR Tm1743 from Thermotoga maritima. Analysis of the Tm1743 crystal structure in complex with tolrestat alone and epalrestat-NADP + shows that tolrestat, but not epalrestat, binding triggers dramatic conformational changes in the anionic site and cofactor binding pocket that prevents accommodation of NADP + . Enzymatic and molecular dynamics simulation analyses further confirm tolrestat as a competitive inhibitor of Tm1743.
Organizational Affiliation:
School of Medicine, Hangzhou Normal University, China.