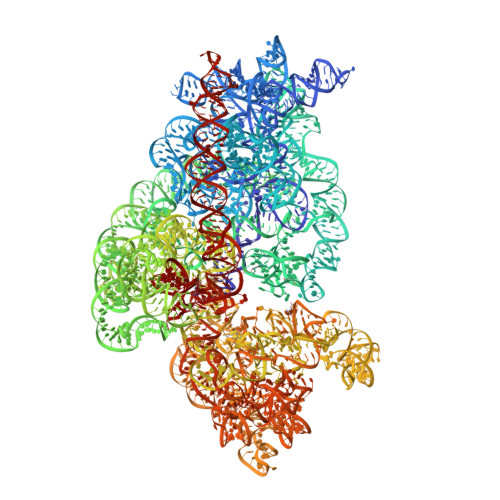
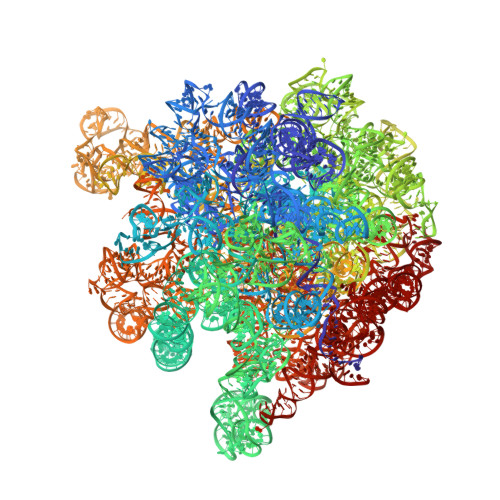
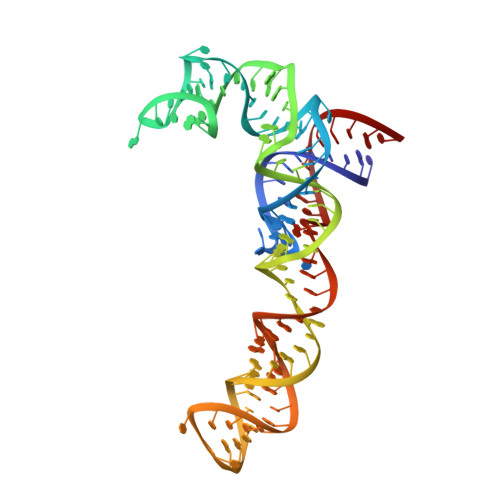
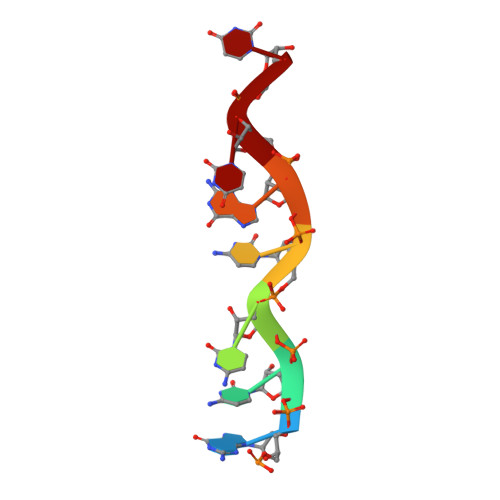
MG Query on MG Download Ideal Coordinates CCD File AC [auth 16S1]
AC [auth 16S1],
MAGNESIUM ION UNX Query on UNX Download Ideal Coordinates CCD File AAA [auth 23S1]
AAA [auth 23S1],
UNKNOWN ATOM OR ION