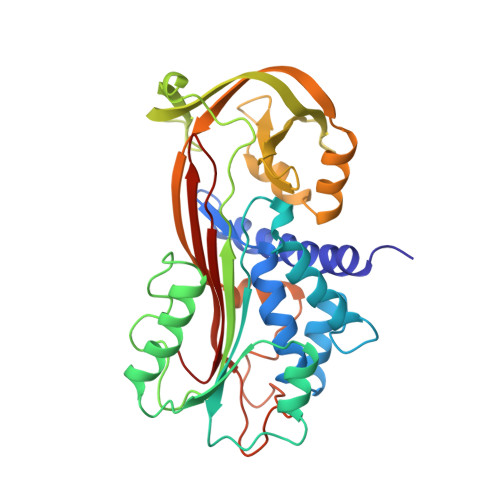
Mechanisms contributing to the conformational and functional flexibility of plasminogen activator inhibitor-1.
Aertgeerts, K., De Bondt, H.L., De Ranter, C.J., Declerck, P.J.(1995) Nat Struct Biol 2: 891-897
- PubMed: 7552714
- DOI: https://doi.org/10.1038/nsb1095-891
- Primary Citation of Related Structures:
9PAI - PubMed Abstract:
Plasminogen activator inhibitor-1 (PAI-1) is unique among the serine proteinase inhibitors (serpins) in that it can adopt at least three different conformations (active, substrate and latent). We report the X-ray structure of a cleaved substrate variant of human PAI-1, which has a new beta-strand s4A formed by insertion of the amino-terminal portion of the reactive-site loop into beta-sheet A subsequent to cleavage. This is in contrast to the previous suggestion that the non-inhibitory function of substrate-type serpins is mainly due to an inability of the reactive-site loop to adopt this conformation. Comparison with the structure of latent PAI-1 provides insights into the molecular determinants responsible for the transition of the stressed active conformation to the thermostable latent conformation.
Organizational Affiliation:
Laboratory for Analytical Chemistry and Medicinal Physicochemistry, Faculty of Pharmaceutical Sciences, Katholieke Universiteit Leuven, Belgium.