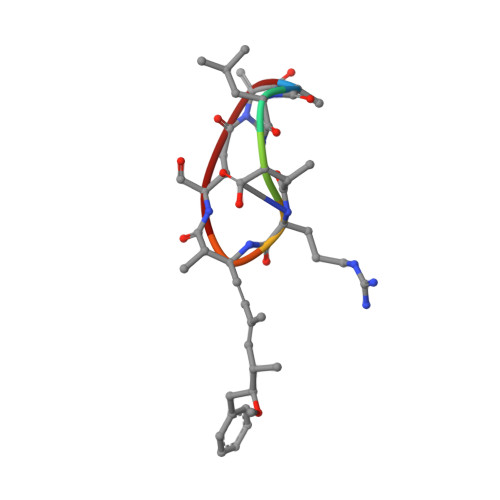
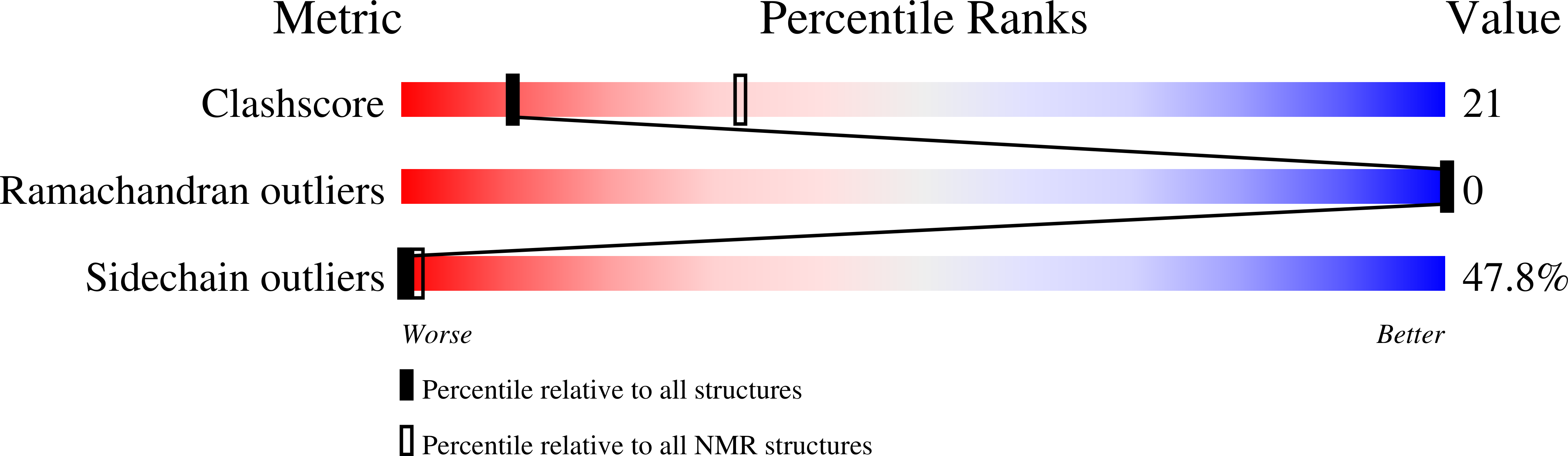Comparison of the solution structures of microcystin-LR and motuporin.
Bagu, J.R., Sonnichsen, F.D., Williams, D., Andersen, R.J., Sykes, B.D., Holmes, C.F.(1995) Nat Struct Biol 2: 114-116
- PubMed: 7749913
- DOI: https://doi.org/10.1038/nsb0295-114
- Primary Citation of Related Structures:
1EVA, 1EVB, 1EVC, 1EVD - PubMed Abstract:
A comparison of the structures of two cyanobacterial toxins yields insights into how they may inhibit protein phosphatase-1 and -2A and why microcystins but not motuporin may covalently modify their protein phosphatase targets.