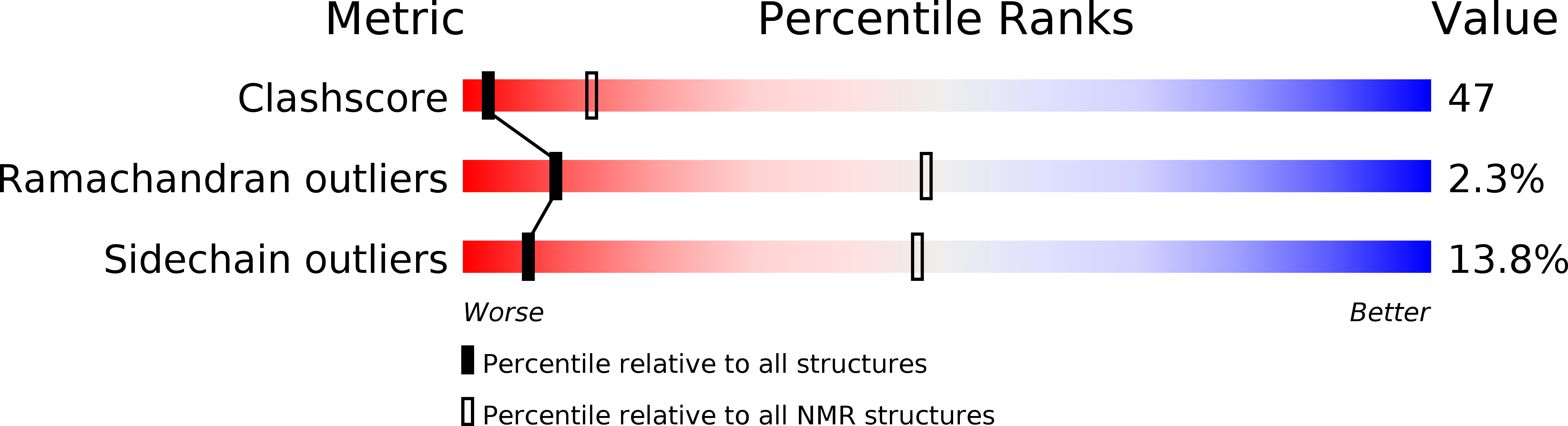The v-myc-induced Q83 lipocalin is a siderocalin.
Coudevylle, N., Geist, L., Hotzinger, M., Hartl, M., Kontaxis, G., Bister, K., Konrat, R.(2010) J Biol Chem 285: 41646-41652
- PubMed: 20826777
- DOI: https://doi.org/10.1074/jbc.M110.123331
- Primary Citation of Related Structures:
2KT4 - PubMed Abstract:
Siderocalins are atypical lipocalins able to capture siderophores with high affinity. They contribute to the innate immune response by interfering with bacterial siderophore-mediated iron uptake but are also involved in numerous physiological processes such as inflammation, iron delivery, tissue differentiation, and cancer progression. The Q83 lipocalin was originally identified based on its overexpression in quail embryo fibroblasts transformed by the v-myc oncogene. We show here that Q83 is a siderocalin, binding the siderophore enterobactin with an affinity and mode of binding nearly identical to that of neutrophil gelatinase-associated lipocalin (NGAL), the prototypical siderocalin. This strengthens the role of siderocalins in cancer progression and inflammation. In addition, we also present the solution structure of Q83 in complex with intact enterobactin and a detailed analysis of the Q83 binding mode, including mutagenesis of the critical residues involved in enterobactin binding. These data provide a first insight into the molecular details of siderophore binding and delineate the common molecular properties defining the siderocalin protein family.
Organizational Affiliation:
Department of Structural and Computational Biology, Max F Perutz Laboratories, University of Vienna, 1030 Vienna, Austria. nicolas.coudevylle@univie.ac.at