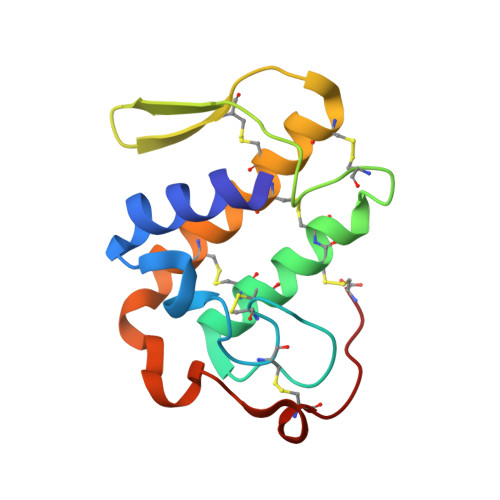
Crystal structure of the complex formed between group II phospholipase A2 and anti-inflammatory agent 2-[(2,6-Dichlorophenyl)amino] benzeneacetic acid at 2.7A resolution
Senthil kumar, R., Singh, N., Ethayathulla, A.S., Prem kumar, R., Sharma, S., Singh, T.P.To be published.