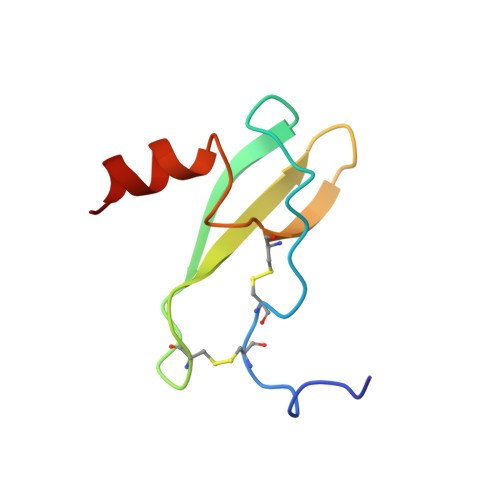
Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
Crump, M.P., Gong, J.H., Loetscher, P., Rajarathnam, K., Amara, A., Arenzana-Seisdedos, F., Virelizier, J.L., Baggiolini, M., Sykes, B.D., Clark-Lewis, I.(1997) EMBO J 16: 6996-7007
- PubMed: 9384579
- DOI: https://doi.org/10.1093/emboj/16.23.6996
- Primary Citation of Related Structures:
1SDF, 2SDF - PubMed Abstract:
The three-dimensional structure of stromal cell-derived factor-1 (SDF-1) was determined by NMR spectroscopy. SDF-1 is a monomer with a disordered N-terminal region (residues 1-8), and differs from other chemokines in the packing of the hydrophobic core and surface charge distribution. Results with analogs showed that the N-terminal eight residues formed an important receptor binding site; however, only Lys-1 and Pro-2 were directly involved in receptor activation. Modification to Lys-1 and/or Pro-2 resulted in loss of activity, but generated potent SDF-1 antagonists. Residues 12-17 of the loop region, which we term the RFFESH motif, unlike the N-terminal region, were well defined in the SDF-1 structure. The RFFESH formed a receptor binding site, which we propose to be an important initial docking site of SDF-1 with its receptor. The ability of the SDF-1 analogs to block HIV-1 entry via CXCR4, which is a HIV-1 coreceptor for the virus in addition to being the receptor for SDF-1, correlated with their affinity for CXCR4. Activation of the receptor is not required for HIV-1 inhibition.
- Protein Engineering Network of Centers of Excellence (PENCE) and Department of Biochemistry, 713 Heritage Medical Research Center, University of Alberta, Edmonton, Alberta, Canada T6G 2S2.
Organizational Affiliation: