Comparative Structural, Kinetic and Inhibitor Studies of Trypanosoma Brucei Trypanothione Reductase with T. Cruzi.
Jones, D., Ariza, A., Chow, W.H.A., Oza, S.L., Fairlamb, A.H.(2010) Mol Biochem Parasitol 169: 12
- PubMed: 19747949
- DOI: https://doi.org/10.1016/j.molbiopara.2009.09.002
- Primary Citation of Related Structures:
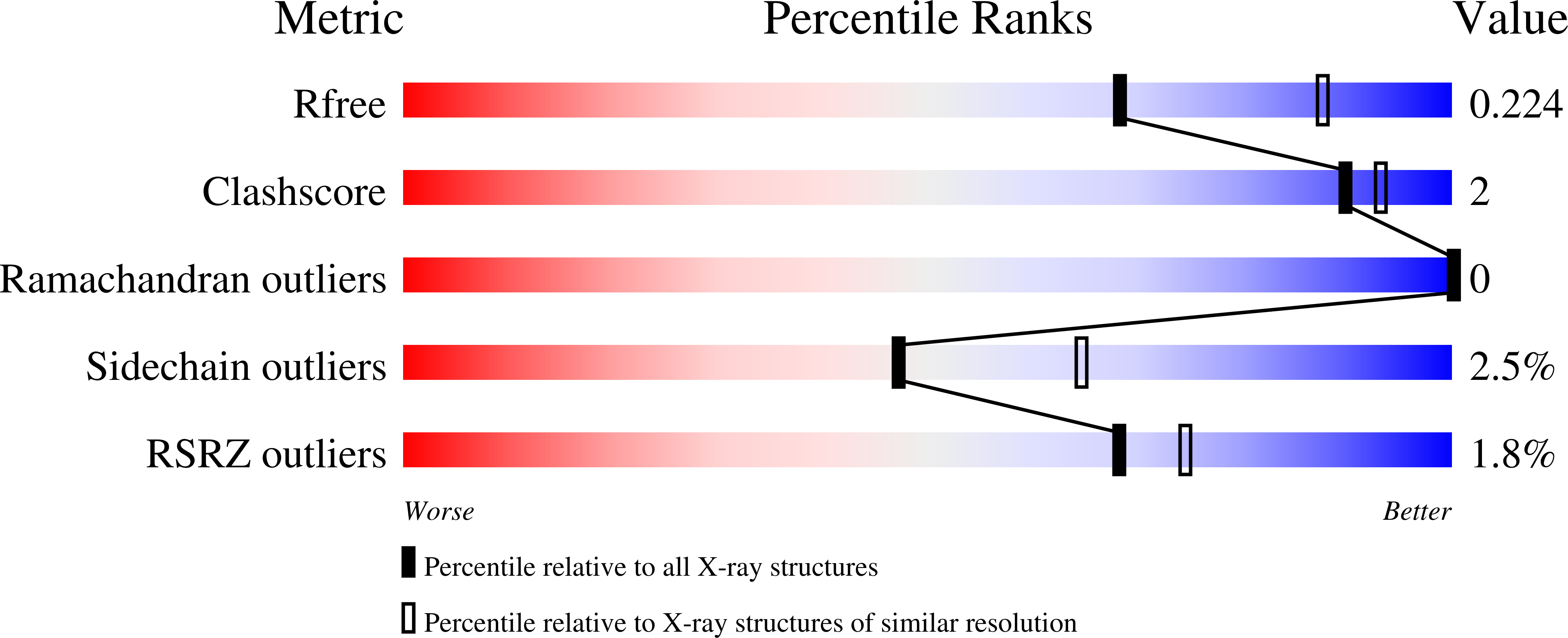
2WBA - PubMed Abstract:
As part of a drug discovery programme to discover new treatments for human African trypanosomiasis, recombinant trypanothione reductase from Trypanosoma brucei has been expressed, purified and characterized. The crystal structure was solved by molecular replacement to a resolution of 2.3A and found to be nearly identical to the T. cruzi enzyme (root mean square deviation 0.6A over 482 Calpha atoms). Kinetically, the K(m) for trypanothione disulphide for the T. brucei enzyme was 4.4-fold lower than for T. cruzi measured by either direct (NADPH oxidation) or DTNB-coupled assay. The K(m) for NADPH for the T. brucei enzyme was found to be 0.77microM using an NADPH-regenerating system coupled to reduction of DTNB. Both enzymes were assayed for inhibition at their respective S=K(m) values for trypanothione disulphide using a range of chemotypes, including CNS-active drugs such as clomipramine, trifluoperazine, thioridazine and citalopram. The relative IC(50) values for the two enzymes were found to vary by no more than 3-fold. Thus trypanothione reductases from these species are highly similar in all aspects, indicating that they may be used interchangeably for structure-based inhibitor design and high-throughput screening.
Organizational Affiliation:
The Wellcome Trust Biocentre, University of Dundee, Scotland, UK.