Extending the Usability of the Phasing Power of Diselenide Bonds: Secys Sad Phasing of Csgc Using a Non-Auxotrophic Strain.
Salgado, P.S., Taylor, J.D., Cota, E., Matthews, S.J.(2011) Acta Crystallogr D Biol Crystallogr 67: 8
- PubMed: 21206057
- DOI: https://doi.org/10.1107/S0907444910042022
- Primary Citation of Related Structures:
2XSK - PubMed Abstract:
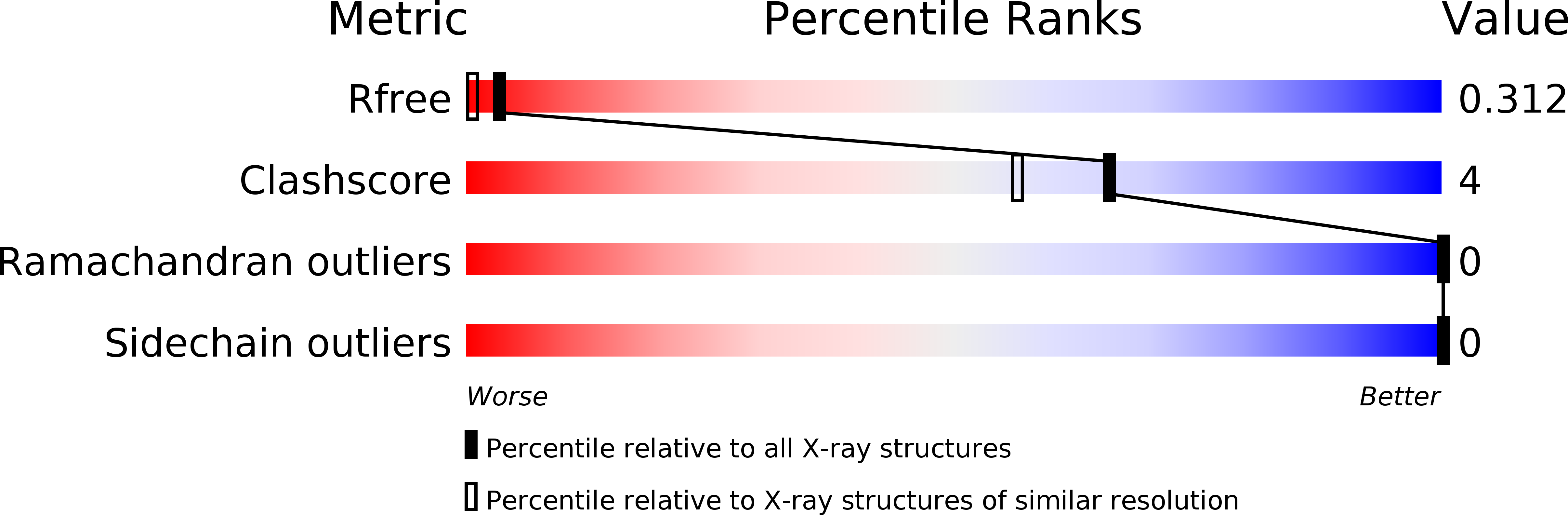
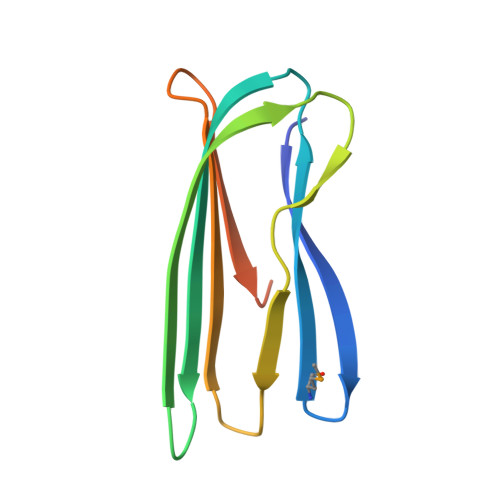
The CsgC protein is a component of the curli system in Escherichia coli. Reported here is the successful incorporation of selenocysteine (SeCys) and selenomethionine (SeMet) into recombinant CsgC, yielding derivatized crystals suitable for structural determination. Unlike in previous reports, a standard autotrophic expression strain was used and only single-wavelength anomalous dispersion (SAD) data were required for successful phasing. The level of SeCys/SeMet incorporation was estimated by mass spectrometry to be about 80%. The native protein crystallized in two different crystal forms (form 1 belonging to space group C222(1) and form 2 belonging to space group C2), which diffracted to 2.4 and 2.0 Å resolution, respectively, whilst Se-derivatized protein crystallized in space group C2 and diffracted to 1.7 Å resolution. The Se-derivatized crystals are suitable for SAD structure determination using only the anomalous signal derived from the SeCys residues. These results extend the usability of SeCys labelling to more general and less favourable cases, rendering it a suitable alternative to traditional phasing approaches.
Organizational Affiliation:
Imperial College London, England.