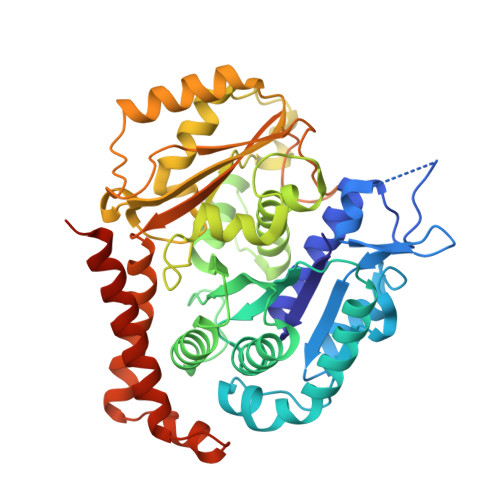
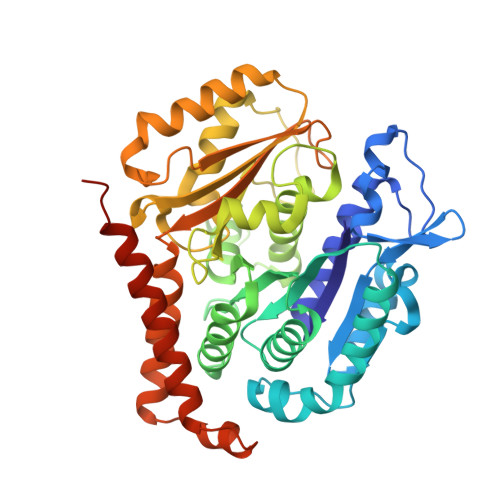
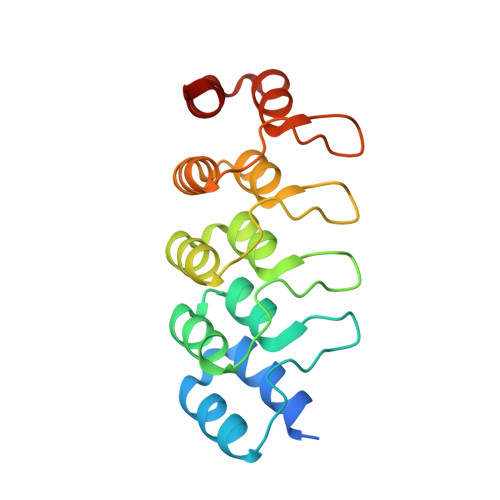
Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length
Zheng, X., Ramani, A., Soni, K., Gottardo, M., Zheng, S., Ming Gooi, L., Li, W., Feng, S., Mariappan, A., Wason, A., Widlund, P., Pozniakovsky, A., Poser, I., Deng, H., Ou, G., Riparbelli, M., Giuliano, C., Hyman, A.A., Sattler, M., Gopalakrishnan, J., Li, H.(2016) Nat Commun 7: 11874-11874
- PubMed: 27306797
- DOI: https://doi.org/10.1038/ncomms11874
- Primary Citation of Related Structures:
5EIB - PubMed Abstract:
Centrioles and cilia are microtubule-based structures, whose precise formation requires controlled cytoplasmic tubulin incorporation. How cytoplasmic tubulin is recognized for centriolar/ciliary-microtubule construction remains poorly understood. Centrosomal-P4.1-associated-protein (CPAP) binds tubulin via its PN2-3 domain. Here, we show that a C-terminal loop-helix in PN2-3 targets β-tubulin at the microtubule outer surface, while an N-terminal helical motif caps microtubule's α-β surface of β-tubulin. Through this, PN2-3 forms a high-affinity complex with GTP-tubulin, crucial for defining numbers and lengths of centriolar/ciliary-microtubules. Surprisingly, two distinct mutations in PN2-3 exhibit opposite effects on centriolar/ciliary-microtubule lengths. CPAP(F375A), with strongly reduced tubulin interaction, causes shorter centrioles and cilia exhibiting doublet- instead of triplet-microtubules. CPAP(EE343RR) that unmasks the β-tubulin polymerization surface displays slightly reduced tubulin-binding affinity inducing over-elongation of newly forming centriolar/ciliary-microtubules by enhanced dynamic release of its bound tubulin. Thus CPAP regulates delivery of its bound-tubulin to define the size of microtubule-based cellular structures using a 'clutch-like' mechanism.
- Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: