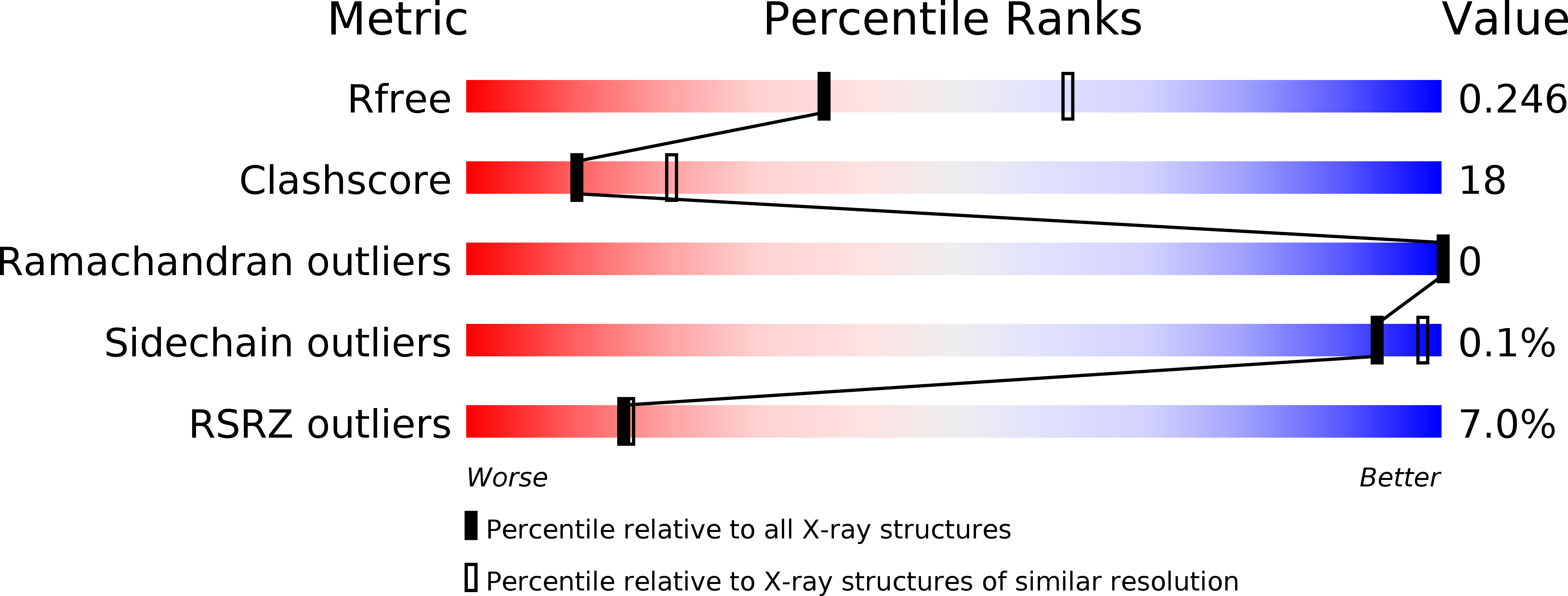
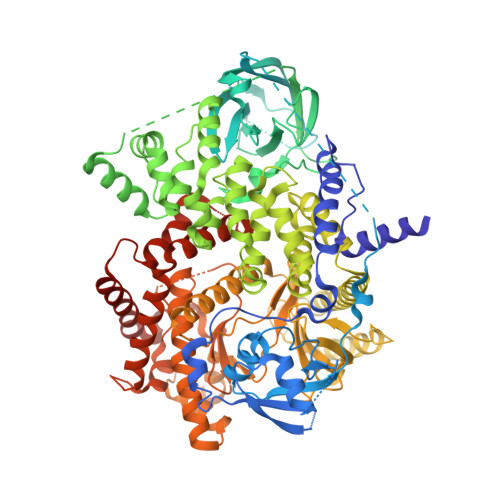
Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Bohnacker, T., Prota, A.E., Beaufils, F., Burke, J.E., Melone, A., Inglis, A.J., Rageot, D., Sele, A.M., Cmiljanovic, V., Cmiljanovic, N., Bargsten, K., Aher, A., Akhmanova, A., Diaz, J.F., Fabbro, D., Zvelebil, M., Williams, R.L., Steinmetz, M.O., Wymann, M.P.(2017) Nat Commun 8: 14683-14683
- PubMed: 28276440
- DOI: https://doi.org/10.1038/ncomms14683
- Primary Citation of Related Structures:
5JHA, 5JHB, 5M7E, 5M7G, 5M8D, 5M8G - PubMed Abstract:
BKM120 (Buparlisib) is one of the most advanced phosphoinositide 3-kinase (PI3K) inhibitors for the treatment of cancer, but it interferes as an off-target effect with microtubule polymerization. Here, we developed two chemical derivatives that differ from BKM120 by only one atom. We show that these minute changes separate the dual activity of BKM120 into discrete PI3K and tubulin inhibitors. Analysis of the compounds cellular growth arrest phenotypes and microtubule dynamics suggest that the antiproliferative activity of BKM120 is mainly due to microtubule-dependent cytotoxicity rather than through inhibition of PI3K. Crystal structures of BKM120 and derivatives in complex with tubulin and PI3K provide insights into the selective mode of action of this class of drugs. Our results raise concerns over BKM120's generally accepted mode of action, and provide a unique mechanistic basis for next-generation PI3K inhibitors with improved safety profiles and flexibility for use in combination therapies.
Organizational Affiliation:
Department of Biomedicine, University of Basel, 4058 Basel, Switzerland.