Structural Basis for Gating and Activation of RyR1.
des Georges, A., Clarke, O.B., Zalk, R., Yuan, Q., Condon, K.J., Grassucci, R.A., Hendrickson, W.A., Marks, A.R., Frank, J.(2016) Cell 167: 145-157.e17
- PubMed: 27662087
- DOI: https://doi.org/10.1016/j.cell.2016.08.075
- Primary Citation of Related Structures:
5T15, 5T9M, 5T9N, 5T9R, 5T9S, 5T9V, 5TA3, 5TAL, 5TAM, 5TAN, 5TAP, 5TAQ, 5TAS, 5TAT, 5TAU, 5TAV, 5TAW, 5TAX, 5TAY, 5TAZ, 5TB0, 5TB1, 5TB2, 5TB3, 5TB4 - PubMed Abstract:
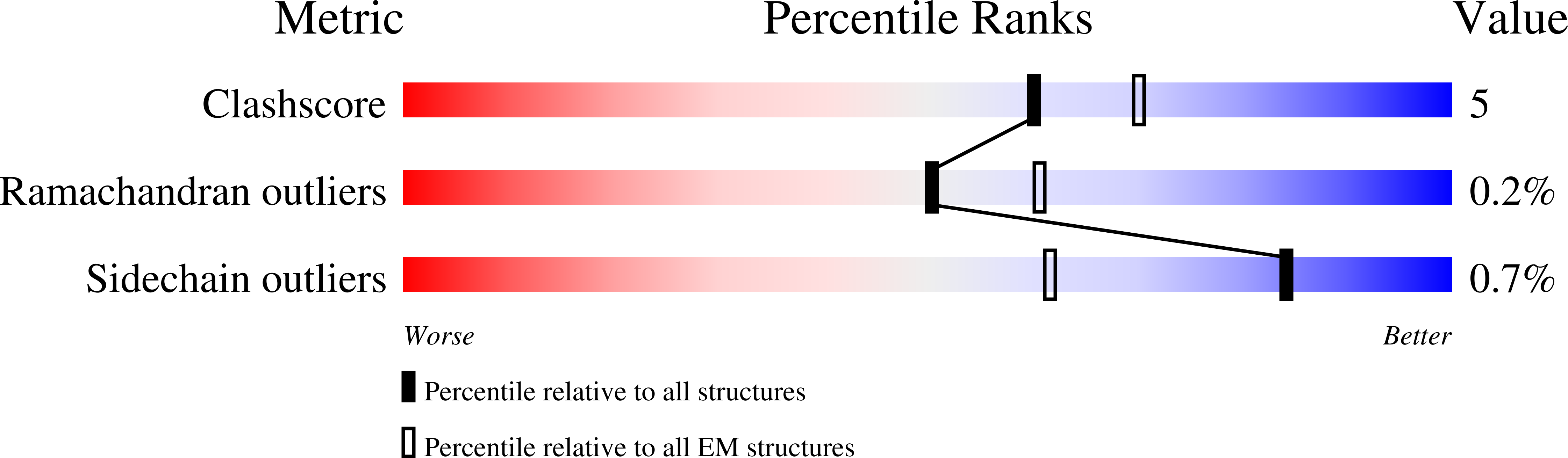
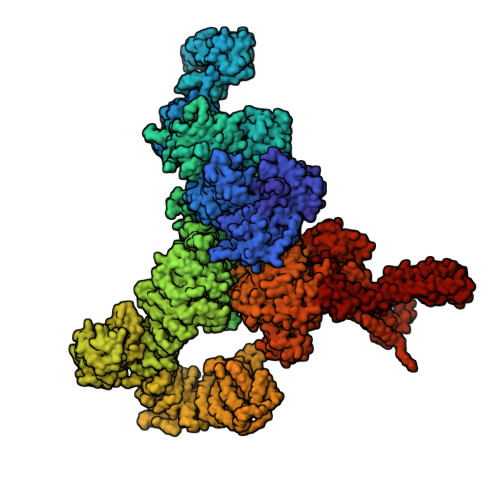
The type-1 ryanodine receptor (RyR1) is an intracellular calcium (Ca(2+)) release channel required for skeletal muscle contraction. Here, we present cryo-EM reconstructions of RyR1 in multiple functional states revealing the structural basis of channel gating and ligand-dependent activation. Binding sites for the channel activators Ca(2+), ATP, and caffeine were identified at interdomain interfaces of the C-terminal domain. Either ATP or Ca(2+) alone induces conformational changes in the cytoplasmic assembly ("priming"), without pore dilation. In contrast, in the presence of all three activating ligands, high-resolution reconstructions of open and closed states of RyR1 were obtained from the same sample, enabling analyses of conformational changes associated with gating. Gating involves global conformational changes in the cytosolic assembly accompanied by local changes in the transmembrane domain, which include bending of the S6 transmembrane segment and consequent pore dilation, displacement, and deformation of the S4-S5 linker and conformational changes in the pseudo-voltage-sensor domain.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA; Howard Hughes Medical Institute, Columbia University, New York, NY 10032, USA.