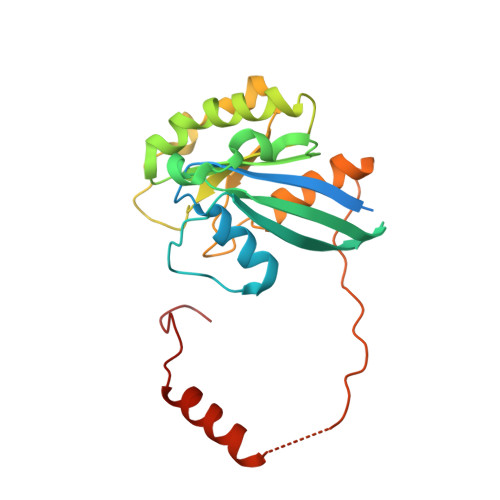
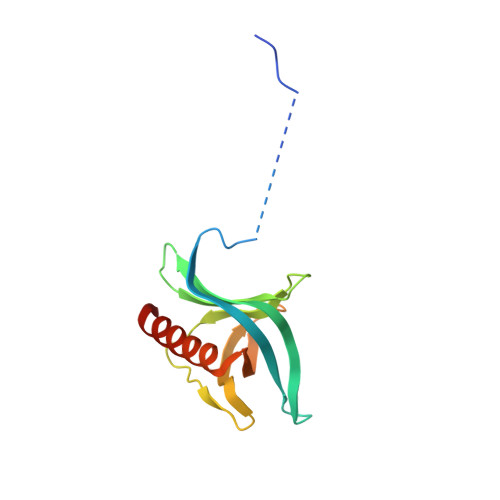
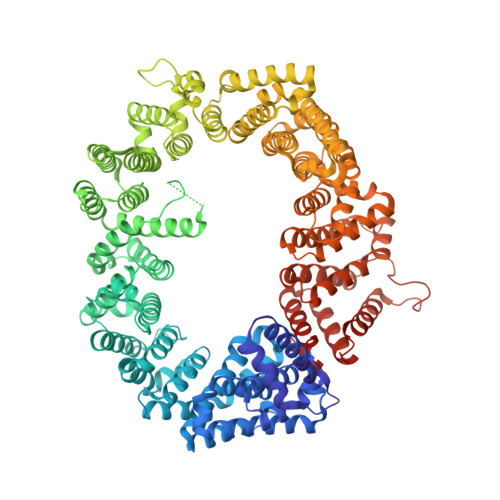
Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals.
Fung, H.Y., Fu, S.C., Chook, Y.M.(2017) Elife 6
- PubMed: 28282025
- DOI: https://doi.org/10.7554/eLife.23961
- Primary Citation of Related Structures:
5UWH, 5UWI, 5UWJ, 5UWO, 5UWP, 5UWQ, 5UWR, 5UWS, 5UWT, 5UWU, 5UWW - PubMed Abstract:
Nuclear export receptor CRM1 binds highly variable nuclear export signals (NESs) in hundreds of different cargoes. Previously we have shown that CRM1 binds NESs in both polypeptide orientations (Fung et al., 2015). Here, we show crystal structures of CRM1 bound to eight additional NESs which reveal diverse conformations that range from loop-like to all-helix, which occupy different extents of the invariant NES-binding groove. Analysis of all NES structures show 5-6 distinct backbone conformations where the only conserved secondary structural element is one turn of helix that binds the central portion of the CRM1 groove. All NESs also participate in main chain hydrogen bonding with human CRM1 Lys568 side chain, which acts as a specificity filter that prevents binding of non-NES peptides. The large conformational range of NES backbones explains the lack of a fixed pattern for its 3-5 hydrophobic anchor residues, which in turn explains the large array of peptide sequences that can function as NESs.
Organizational Affiliation:
Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, United States.