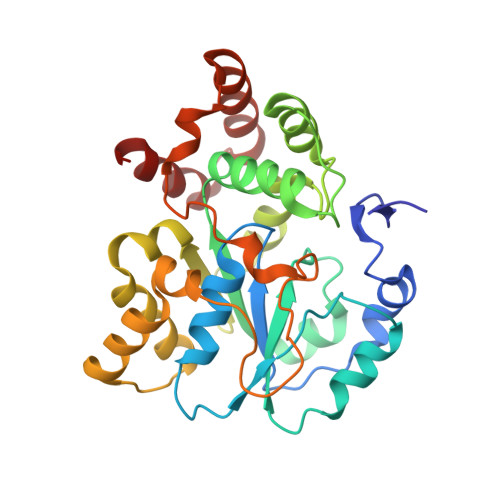
A new type of sulfation reaction: C -sulfonation for alpha , beta-unsaturated carbonyl groups by a novel sulfotransferase SULT7A1.
Kurogi, K., Sakakibara, Y., Hashiguchi, T., Kakuta, Y., Kanekiyo, M., Teramoto, T., Fukushima, T., Bamba, T., Matsumoto, J., Fukusaki, E., Kataoka, H., Suiko, M.(2024) PNAS Nexus 3: pgae097-pgae097
- PubMed: 38487162
- DOI: https://doi.org/10.1093/pnasnexus/pgae097
- Primary Citation of Related Structures:
5X2B - PubMed Abstract:
Cytosolic sulfotransferases (SULTs) are cytosolic enzymes that catalyze the transfer of sulfonate group to key endogenous compounds, altering the physiological functions of their substrates. SULT enzymes catalyze the O -sulfonation of hydroxy groups or N -sulfonation of amino groups of substrate compounds. In this study, we report the discovery of C -sulfonation of α,β-unsaturated carbonyl groups mediated by a new SULT enzyme, SULT7A1, and human SULT1C4. Enzymatic assays revealed that SULT7A1 is capable of transferring the sulfonate group from 3'-phosphoadenosine 5'-phosphosulfate to the α-carbon of α,β-unsaturated carbonyl-containing compounds, including cyclopentenone prostaglandins as representative endogenous substrates. Structural analyses of SULT7A1 suggest that the C -sulfonation reaction is catalyzed by a novel mechanism mediated by His and Cys residues in the active site. Ligand-activity assays demonstrated that sulfonated 15-deoxy prostaglandin J 2 exhibits antagonist activity against the prostaglandin receptor EP2 and the prostacyclin receptor IP. Modification of α,β-unsaturated carbonyl groups via the new prostaglandin-sulfonating enzyme, SULT7A1, may regulate the physiological function of prostaglandins in the gut. Discovery of C -sulfonation of α,β-unsaturated carbonyl groups will broaden the spectrum of potential substrates and physiological functions of SULTs.
- Department of Biochemistry and Applied Biosciences, Faculty of Agriculture, University of Miyazaki, Miyazaki 889-2192, Japan.
Organizational Affiliation: