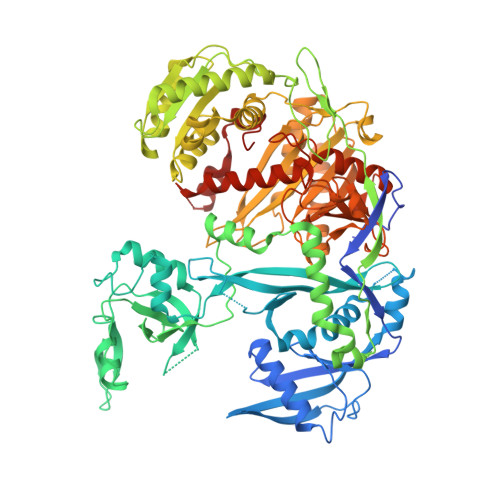
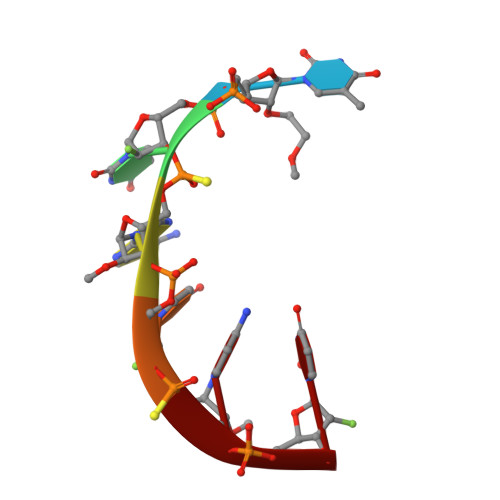Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide.
Schirle, N.T., Kinberger, G.A., Murray, H.F., Lima, W.F., Prakash, T.P., MacRae, I.J.(2016) J Am Chem Soc 138: 8694-8697
- PubMed: 27380263
- DOI: https://doi.org/10.1021/jacs.6b04454
- Primary Citation of Related Structures:
5JS1, 5JS2 - PubMed Abstract:
Incorporation of chemical modifications into small interfering RNAs (siRNAs) increases their metabolic stability and improves their tissue distribution. However, how these modifications impact interactions with Argonaute-2 (Ago2), the molecular target of siRNAs, is not known. Herein we present the crystal structure of human Ago2 bound to a metabolically stable siRNA containing extensive backbone modifications. Comparison to the structure of an equivalent unmodified-siRNA complex indicates that the structure of Ago2 is relatively unaffected by chemical modifications in the bound siRNA. In contrast, the modified siRNA appears to be much more plastic and shifts, relative to the unmodified siRNA, to optimize contacts with Ago2. Structure-activity analysis reveals that even major conformational perturbations in the 3' half of the siRNA seed region have a relatively modest effect on knockdown potency. These findings provide an explanation for a variety of modification patterns tolerated in siRNAs and a structural basis for advancing therapeutic siRNA design.
- Department of Integrative Computational and Structural Biology, The Scripps Research Institute , La Jolla, California 92037, United States.
Organizational Affiliation: