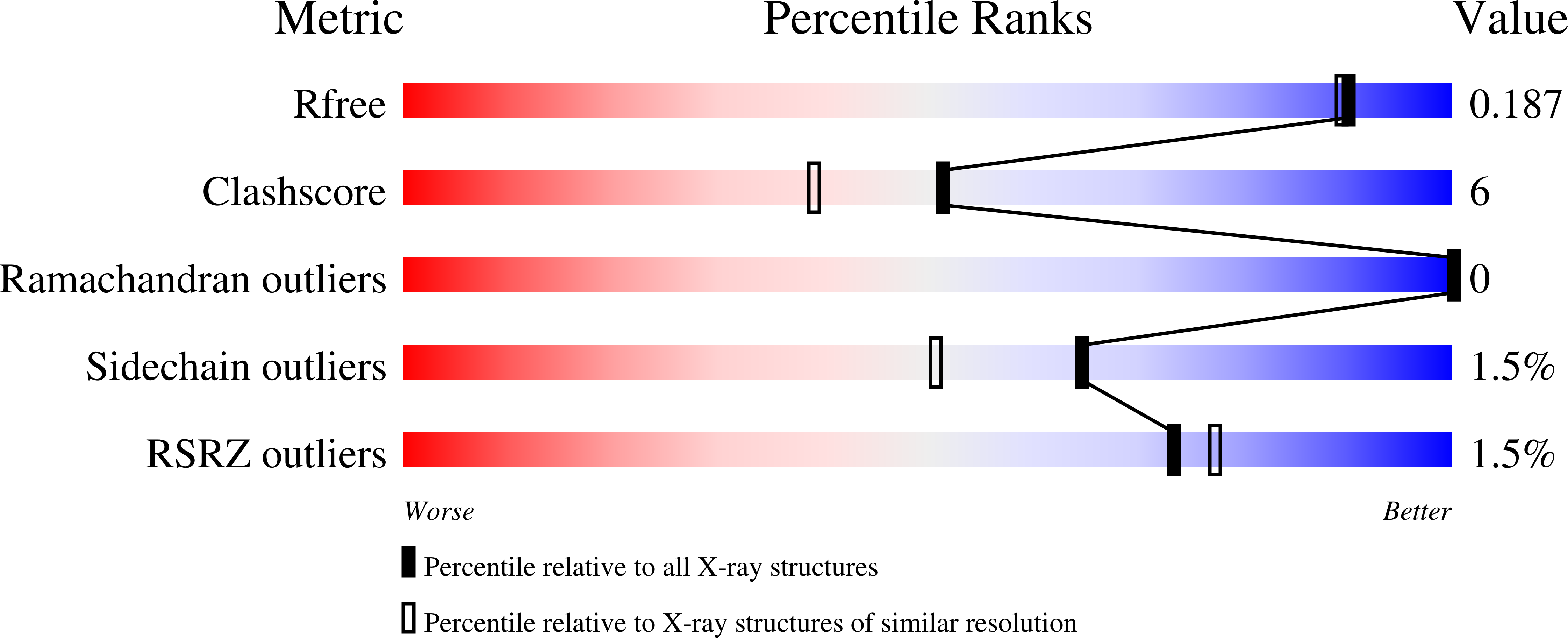
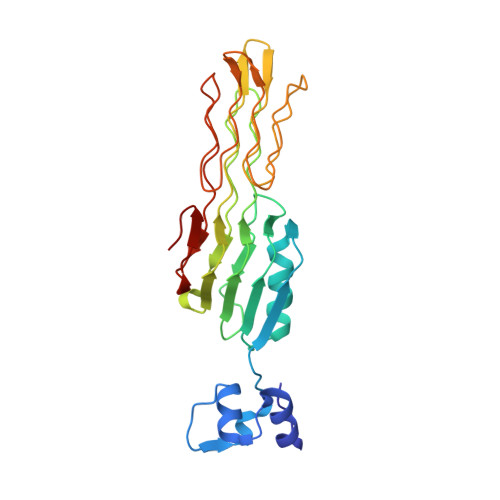
Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition.
Dunne, M., Denyes, J.M., Arndt, H., Loessner, M.J., Leiman, P.G., Klumpp, J.(2018) Structure 26: 1573-1582.e4
- PubMed: 30244968
- DOI: https://doi.org/10.1016/j.str.2018.07.017
- Primary Citation of Related Structures:
6F45 - PubMed Abstract:
The ability of phages to infect specific bacteria has led to their exploitation as bio-tools for bacterial remediation and detection. Many phages recognize bacterial hosts via adhesin tips of their long tail fibers (LTFs). Adhesin sequence plasticity modulates receptor specificity, and thus primarily defines a phage's host range. Here we present the crystal structure of an adhesin (gp38) attached to a trimeric β-helical tip (gp37) from the Salmonella phage S16 LTF. Gp38 contains rare polyglycine type II helices folded into a packed lattice, herein designated "PG II sandwich." Sequence variability within the domain is limited to surface-exposed helices and distal loops that form putative receptor-binding sites. In silico analyses revealed a prevalence of the adhesin architecture among T-even phages, excluding the archetypal T4 phage. Overall, S16 LTF provides a valuable model for understanding binding mechanisms of phage adhesins, and for engineering of phage adhesins with expandable or modulated host ranges.
Organizational Affiliation:
Institute of Food, Nutrition and Health, ETH Zurich, Zurich, Switzerland. Electronic address: matthew.dunne@hest.ethz.ch.