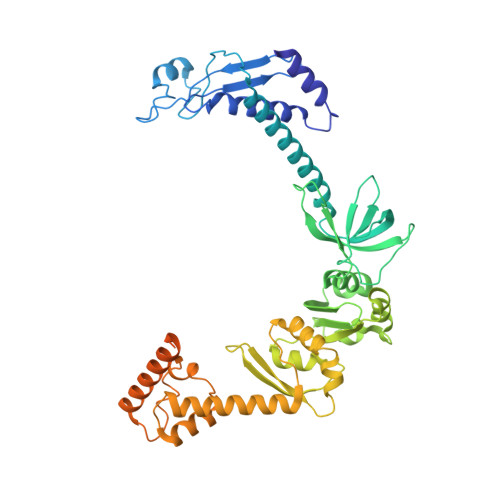
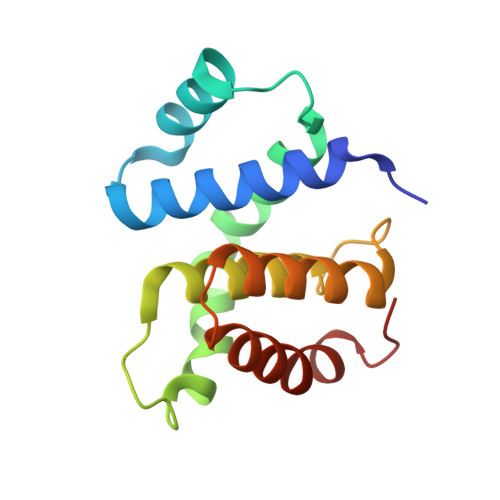
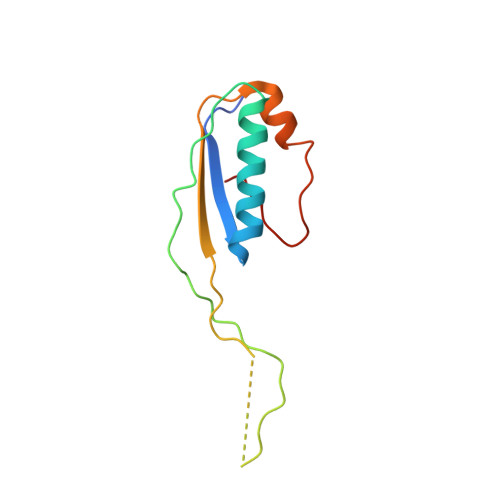
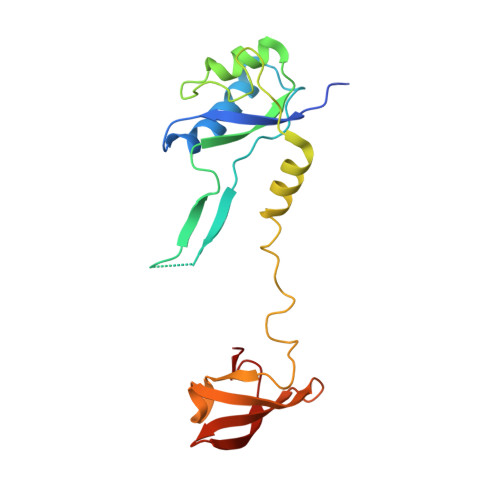
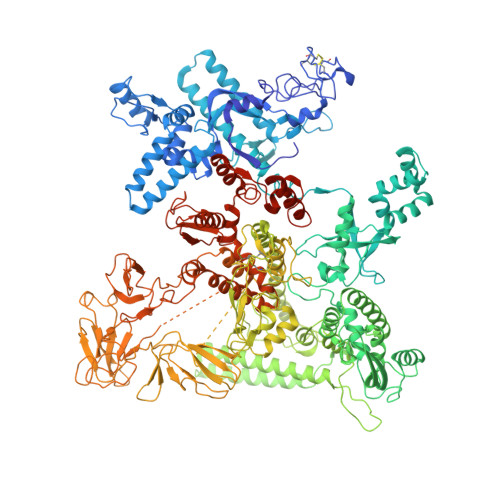
Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor.
Krupp, F., Said, N., Huang, Y.H., Loll, B., Burger, J., Mielke, T., Spahn, C.M.T., Wahl, M.C.(2019) Mol Cell 74: 143-157.e5
- PubMed: 30795892
- DOI: https://doi.org/10.1016/j.molcel.2019.01.016
- Primary Citation of Related Structures:
6GOV - PubMed Abstract:
Bacteriophage λN protein, a model anti-termination factor, binds nascent RNA and host Nus factors, rendering RNA polymerase resistant to all pause and termination signals. A 3.7-Å-resolution cryo-electron microscopy structure and structure-informed functional analyses reveal a multi-pronged strategy by which the intrinsically unstructured λN directly modifies RNA polymerase interactions with the nucleic acids and subverts essential functions of NusA, NusE, and NusG to reprogram the transcriptional apparatus. λN repositions NusA and remodels the β subunit flap tip, which likely precludes folding of pause or termination RNA hairpins in the exit tunnel and disrupts termination-supporting interactions of the α subunit C-terminal domains. λN invades and traverses the RNA polymerase hybrid cavity, likely stabilizing the hybrid and impeding pause- or termination-related conformational changes of polymerase. λN also lines upstream DNA, seemingly reinforcing anti-backtracking and anti-swiveling by NusG. Moreover, λN-repositioned NusA and NusE sequester the NusG C-terminal domain, counteracting ρ-dependent termination. Other anti-terminators likely utilize similar mechanisms to enable processive transcription.
Organizational Affiliation:
Medizinische Physik und Biophysik, Charité - Universitätsmedizin Berlin, Charitéplatz 1, 10117 Berlin, Germany.